lacZ:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
lacZ |
|---|---|
| Gene Synonym(s) |
ECK0341, b0344, JW0335[1], JW0335 |
| Product Desc. |
Component of β-galactosidase[2][3] beta-D-Galactosidase[4] |
| Product Synonyms(s) |
beta-D-galactosidase[1], B0344[2][1], LacZ[2][1], beta-D-galactosidase[2][1], lactase[2][1], beta-D-galactoside galactohydrolase[2][1] , ECK0341, JW0335, b0344 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype |
unable to grow on lactose as a carbon source |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Repressed during biofilm formation. LacZ has an Ig-like beta-sandwich domain (residues 221-334 align to PFAM: PF00703). ebgA, uidA and lacZ are paralogs. Binds TrxA (Kumar, 2004)[5].[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
lacZ |
|---|---|
| Mnemonic |
Lactose |
| Synonyms |
ECK0341, b0344, JW0335[1], JW0335 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
7.81 minutes |
MG1655: 365529..362455 |
||
|
NC_012967: 338861..335787 |
||||
|
NC_012759: 4175805..4179047 |
||||
|
W3110 |
|
W3110: 365529..362455 |
||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
362458 |
Edman degradation |
PMID:97295 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
lacZD202F |
D202F |
Obliterates all binding and catalysis |
seeded from UniProt:P00722 | ||||
|
lacZD202E,N |
D202E,N |
Causes a significant decrease in binding affinity in the absence of monovalent cations or in the presence of potassium ions, but only a moderate decrease in the presence of sodium ions |
seeded from UniProt:P00722 | ||||
|
lacZW1000F,G,L,T |
W1000F,G,L,T |
Decreases affinity for substrate |
seeded from UniProt:P00722 | ||||
|
lacZF602A |
F602A |
Decreases the stability of the loop 794-804 |
seeded from UniProt:P00722 | ||||
|
lacZG795A |
G795A |
It forces the apoenzyme to adopt the closed rather than the open conformation. Reduces the binding affinity |
seeded from UniProt:P00722 | ||||
|
lacZE798A,L |
E798A,L |
The catalytic efficiency is not increased, when the sodium concentration increases |
seeded from UniProt:P00722 | ||||
|
lacZE798D,Q |
E798D,Q |
Small increase of the catalytic efficiency, when the sodium concentration increases |
seeded from UniProt:P00722 | ||||
|
lacZH541E,F,N |
H541E,F,N |
Poorly reactive with galactosyl substrates. Less stable to heat than wild-type |
seeded from UniProt:P00722 | ||||
|
lacZE538Q |
E538Q |
10000-fold decrease in the beta- galactosidase activity |
seeded from UniProt:P00722 | ||||
|
lacZE462H |
E462H |
Slowly inactivates galactosidase activity by reducing the binding of magnesium. It increases binding specificity |
seeded from UniProt:P00722 | ||||
|
lacZH358D,F,L,N |
H358D,F,L,N |
Less stable to heat than wild-type. Causes significant destabilizations of the first transition state |
seeded from UniProt:P00722 | ||||
|
lacZH392E,F,K |
H392E,F,K |
Essentially inactive unless very rapid purification. Causes very large destabilizations of the transition state |
seeded from UniProt:P00722 | ||||
|
lacZ4 |
|||||||
|
lacZ43(Fs) |
frameshift mutation | ||||||
|
lacZ53(Am) |
amber (UAG) mutation | ||||||
|
lacZ36 |
|||||||
|
lacZ125(Am) |
PMID:5327654 |
amber (UAG) mutation | |||||
|
lacZ73 |
|||||||
|
lacZ46 |
|||||||
|
lacZ827(UGA) |
opal (UGA) mutation | ||||||
|
lacZ82(Am) |
amber (UAG) mutation | ||||||
|
lacZ39(del) |
|||||||
|
lacZ13(Oc) |
ochre (UAA) mutation | ||||||
|
lacZ90(Oc) |
PMID:6790520 PMID:5327654 |
ochre (UAA) mutation | |||||
|
lacZ625(Am) |
PMID:5327654 |
amber (UAG) mutation | |||||
|
lacZ118(Oc) |
PMID:5327654 PMID:412841 PMID:4876923 |
ochre (UAA) mutation | |||||
|
lacZ659(Oc) |
PMID:5327654 |
ochre (UAA) mutation | |||||
|
lacZ404(Oc) |
PMID:5327654 |
ochre (UAA) mutation | |||||
|
lacZ521(UGA) |
PMID:4915862 |
opal (UGA) mutation | |||||
|
lacZ813(UGA) |
PMID:4915862 |
opal (UGA) mutation | |||||
|
lacZ105(Am) |
amber (UAG) mutation | ||||||
|
lacZ178 |
|||||||
|
lacZ281(Am) |
amber (UAG) mutation | ||||||
|
lacZ57(del) |
|||||||
|
lacZ62 |
|||||||
|
lacZ608(Am) |
PMID:5327654 PMID:4915862 |
amber (UAG) mutation | |||||
|
lacZ84 |
|||||||
|
lacZ75(Fs) |
frameshift mutation | ||||||
|
lacZ482(Am) |
amber (UAG) mutation | ||||||
|
lacZ58(del)(M15) |
|||||||
|
lacZ98::Tn10 |
|||||||
|
lacZ545(Am) |
amber (UAG) mutation | ||||||
|
lacZ2286(Am) |
amber (UAG) mutation | ||||||
|
lacZ624(Am) |
PMID:4915862 |
amber (UAG) mutation | |||||
|
lacZ95(del) |
|||||||
|
lacZ114((delH)) |
|||||||
|
lacZ55 |
|||||||
|
lacZ2210(Am) |
amber (UAG) mutation | ||||||
|
lacZ332(Fs) |
PMID:1092652 |
||||||
|
lacZ4503(Am) |
amber (UAG) mutation | ||||||
|
lacZ49 |
|||||||
|
lacZ56 |
|||||||
|
lacZ131 |
|||||||
|
lacZ2246 |
|||||||
|
lacZ8(Am) |
PMID:2141650 |
amber (UAG) mutation | |||||
|
lacZ59 |
|||||||
|
lacZ4502::Tn10 |
PMID:337110 |
||||||
|
lacZ4787(del)(::rrnB-3) |
PMID:10829079 |
||||||
|
lacZ4796::Tn5 |
|||||||
|
lacZ536(Am) |
amber (UAG) mutation | ||||||
|
lacZ571(Am) |
PMID:2501784 |
amber (UAG) mutation | |||||
|
lacZ8305::Mu cts62 |
PMID:1111215 |
||||||
|
lacZ4525::Tn10kan |
PMID:9139905 |
||||||
|
lacZ572 |
PMID:2501784 |
||||||
|
lacZ574 |
PMID:2501784 |
||||||
|
lacZ573 |
PMID:2501784 |
||||||
|
lacZ575 |
PMID:2501784 |
||||||
|
lacZ576 |
PMID:2501784 |
||||||
|
lacZ1125 |
PMID:4915862 PMID:5327654 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW0335 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCACCATGATTACGGATTCACT Primer 2:CCTTTTTGACACCAGACCAACTG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: % [7] | ||
|
Linked marker |
est. P1 cotransduction: 87% [7] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10527 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10527 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000520 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0522 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0001183 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
LacZ |
|---|---|
| Synonyms |
beta-D-galactosidase[1], B0344[2][1], LacZ[2][1], beta-D-galactosidase[2][1], lactase[2][1], beta-D-galactoside galactohydrolase[2][1] , ECK0341, JW0335, b0344 |
| Product description |
Component of β-galactosidase[2][3] beta-D-Galactosidase[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000287 |
magnesium ion binding |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_01687 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000287 |
magnesium ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0460 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004565 |
beta-galactosidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004199 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004565 |
beta-galactosidase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:3.2.1.23 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009341 |
beta-galactosidase complex |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004199 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030145 |
manganese ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0464 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030246 |
carbohydrate binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011013 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030246 |
carbohydrate binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR014718 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0031402 |
sodium ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0915 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0043169 |
cation binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013781 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046872 |
metal ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0479 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of β-galactosidase |
could be indirect |
||
|
Protein |
yifB |
PMID:16606699 |
Experiment(s):EBI-1136305 | |
|
Protein |
napC |
PMID:16606699 |
Experiment(s):EBI-1136305 | |
|
Protein |
secA |
PMID:16606699 |
Experiment(s):EBI-1136305 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MTMITDSLAV VLQRRDWENP GVTQLNRLAA HPPFASWRNS EEARTDRPSQ QLRSLNGEWR FAWFPAPEAV PESWLECDLP EADTVVVPSN WQMHGYDAPI YTNVTYPITV NPPFVPTENP TGCYSLTFNV DESWLQEGQT RIIFDGVNSA FHLWCNGRWV GYGQDSRLPS EFDLSAFLRA GENRLAVMVL RWSDGSYLED QDMWRMSGIF RDVSLLHKPT TQISDFHVAT RFNDDFSRAV LEAEVQMCGE LRDYLRVTVS LWQGETQVAS GTAPFGGEII DERGGYADRV TLRLNVENPK LWSAEIPNLY RAVVELHTAD GTLIEAEACD VGFREVRIEN GLLLLNGKPL LIRGVNRHEH HPLHGQVMDE QTMVQDILLM KQNNFNAVRC SHYPNHPLWY TLCDRYGLYV VDEANIETHG MVPMNRLTDD PRWLPAMSER VTRMVQRDRN HPSVIIWSLG NESGHGANHD ALYRWIKSVD PSRPVQYEGG GADTTATDII CPMYARVDED QPFPAVPKWS IKKWLSLPGE TRPLILCEYA HAMGNSLGGF AKYWQAFRQY PRLQGGFVWD WVDQSLIKYD ENGNPWSAYG GDFGDTPNDR QFCMNGLVFA DRTPHPALTE AKHQQQFFQF RLSGQTIEVT SEYLFRHSDN ELLHWMVALD GKPLASGEVP LDVAPQGKQL IELPELPQPE SAGQLWLTVR VVQPNATAWS EAGHISAWQQ WRLAENLSVT LPAASHAIPH LTTSEMDFCI ELGNKRWQFN RQSGFLSQMW IGDKKQLLTP LRDQFTRAPL DNDIGVSEAT RIDPNAWVER WKAAGHYQAE AALLQCTADT LADAVLITTA HAWQHQGKTL FISRKTYRID GSGQMAITVD VEVASDTPHP ARIGLNCQLA QVAERVNWLG LGPQENYPDR LTAACFDRWD LPLSDMYTPY VFPSENGLRC GTRELNYGPH QWRGDFQFNI SRYSQQQLME TSHRHLLHAE EGTWLNIDGF HMGIGGDDSW SPSVSAEFQL SAGRYHYQLV WCQK |
| Length |
1,024 |
| Mol. Wt |
116.482 kDa |
| pI |
5.3 (calculated) |
| Extinction coefficient |
260,690 - 262,690 (calc based on 31 Y, 39 W, and 16 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures
| |||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0001183 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10527 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10527 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000520 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0522 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
|||||||
|
Protein |
E. coli K-12 MG1655 |
31 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
10 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
8a |
molecules/cell/generation |
|
Ribosome Profiling |
Low confidence in the sequencing data set. |
PMID: 24766808 |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
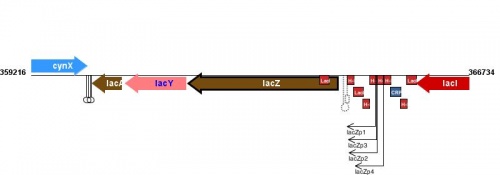 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:365509..365549
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for lacZ | |
|
microarray |
Summary of data for lacZ from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (365438..365669) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
|
GFP Fusion |
Intergenic region (365438..365669) fused to gfpmut2. |
Raw protomer fusion data from Zaslaver, et al.
| ||
| edit table |
<protect></protect>
Notes
Accessions Related to lacZ Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10527 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0522 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10527 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000520 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0001183 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
E. coli O157 |
LACZ |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10527 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10527 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000520 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0522 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0001183 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ Kumar, JK et al. (2004) Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 101 3759-64 PubMed
- ↑ 6.0 6.1 CGSC: The Coli Genetics Stock Center
- ↑ 7.0 7.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ 8.0 8.1 Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories


