rep:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
rep |
|---|---|
| Gene Synonym(s) |
ECK3770, b3778, JW5604, dasC, mbrA, mmrA, dasC?[1][2], mmrA? |
| Product Desc. |
Component of rep helicase, a single-stranded DNA dependent ATPase[3] ATP-dependent DNA helicase Rep, involved in DNA replication; ssDNA-dependent ATPase[4] |
| Product Synonyms(s) |
DNA helicase and single-stranded DNA-dependent ATPase[1], B3778[2][1], MmrA[2][1], MbrA[2][1], DasC[2][1], Rep[2][1] , dasC, ECK3770, JW5604, mbrA, mmrA?, b3778 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
rep |
|---|---|
| Mnemonic |
Replicase |
| Synonyms |
ECK3770, b3778, JW5604, dasC, mbrA, mmrA, dasC?[1][2], mmrA? |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
85.32 minutes |
MG1655: 3958700..3960721 |
||
|
NC_012967: 3922479..3924500 |
||||
|
NC_012759: 3848369..3850390 |
||||
|
W3110 |
|
W3110: 3676004..3673983 |
||
|
DH10B: 4057620..4059641 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
3958700 |
Edman degradation |
PMID:2999067 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
rep(del) (Keio:JW5604) |
deletion |
deletion |
PMID:16738554 |
||||
|
rep::Tn5KAN-I-SceI (FB21451) |
Insertion at nt 508 in Minus orientation |
PMID:15262929 |
contains pKD46 | ||||
|
Δrep::kan |
deletion |
Biolog:respiration |
unable to respire Acetate |
PMID:16095938 |
|||
|
Δrep::kan |
deletion |
Biolog:respiration |
unable to respire a-Hydroxybutyrate |
PMID:16095938 |
|||
|
Δrep::kan |
deletion |
Biolog:respiration |
unable to respire a-Ketobutyrate |
PMID:16095938 |
|||
|
Δrep::kan |
deletion |
Biolog:respiration |
unable to respire a-Ketoglutarate |
PMID:16095938 |
|||
|
Δrep::kan |
deletion |
Biolog:respiration |
unable to respire Succinate |
PMID:16095938 |
|||
|
Δrep::kan |
deletion |
Biolog:respiration |
unable to respire Bromosuccinate |
PMID:16095938 |
|||
|
Δrep::kan |
deletion |
Biolog:respiration |
unable to respire L-Alanine |
PMID:16095938 |
|||
|
Δrep::kan |
deletion |
Biolog:respiration |
unable to respire L-Asparagine |
PMID:16095938 |
|||
|
Δrep::kan |
deletion |
Biolog:respiration |
unable to respire L-Aspartate |
PMID:16095938 |
|||
|
Δrep::kan |
deletion |
Biolog:respiration |
unable to respire L-Serine |
PMID:16095938 |
|||
|
rep-5 |
|||||||
|
rep-3 |
PMID:383240 |
||||||
|
rep-142(ts) |
temperature sensitive |
||||||
|
[rep-71] |
PMID:6286604 |
null allele | |||||
|
rep-729(del)::kan |
PMID:16738554 |
||||||
|
rep in MDS42 |
deletion |
Sensitivity to |
increased sensitivity to Bicyclomycin |
PMID:21183718 |
Fig. 2 | ||
|
mbrA4 |
Growth Phenotype |
no growth in the first few hours at non-permissive conditions but eventually resumed growth, after exposed to LB cells began to elongate LB-Mac- |
PMID:2176633 |
||||
|
mbrA15 |
Growth Phenotype |
temperature sensitive, forms two distinct populations, one more dense and one less dense than wild type |
PMID:2176633 |
||||
|
mbrA4 |
Resistant to |
camphor resistance |
PMID:2176633 PMID:1925019 |
Table 2 and table 1 | |||
|
mrbA15 |
Resistant to |
Resistant to camphor |
PMID:2176633 PMID:1925019 |
table 2 and table 1 | |||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW5604 |
Plasmid clone |
PMID:16769691 Status:cloned as old JW Primer 1:GCCCGTCTAAACCCCGGCCAACA Primer 2:CCTTTCCCTCGTTTTGCCGCCAT | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 83% [6] | ||
|
metEo-3079::Tn10 |
Linked marker |
est. P1 cotransduction: 10% [6] | |
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10837 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10837 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000828 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0830 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012344 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
Rep |
|---|---|
| Synonyms |
DNA helicase and single-stranded DNA-dependent ATPase[1], B3778[2][1], MmrA[2][1], MbrA[2][1], DasC[2][1], Rep[2][1] , dasC, ECK3770, JW5604, mbrA, mmrA?, b3778 |
| Product description |
Component of rep helicase, a single-stranded DNA dependent ATPase[3] ATP-dependent DNA helicase Rep, involved in DNA replication; ssDNA-dependent ATPase[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000212 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0238 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004003 |
ATP-dependent DNA helicase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000212 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004003 |
ATP-dependent DNA helicase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005752 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004386 |
helicase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0347 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000212 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR014016 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR014017 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005752 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006260 |
DNA replication |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0235 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006268 |
DNA unwinding involved in replication |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005752 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006281 |
DNA repair |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000212 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016787 |
hydrolase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR014016 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016787 |
hydrolase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR014017 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016787 |
hydrolase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0378 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of rep helicase, a single-stranded DNA dependent ATPase |
could be indirect |
| |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MRLNPGQQQA VEFVTGPCLV LAGAGSGKTR VITNKIAHLI RGCGYQARHI AAVTFTNKAA REMKERVGQT LGRKEARGLM ISTFHTLGLD IIKREYAALG MKANFSLFDD TDQLALLKEL TEGLIEDDKV LLQQLISTIS NWKNDLKTPS QAAASAIGER DRIFAHCYGL YDAHLKACNV LDFDDLILLP TLLLQRNEEV RKRWQNKIRY LLVDEYQDTN TSQYELVKLL VGSRARFTVV GDDDQSIYSW RGARPQNLVL LSQDFPALKV IKLEQNYRSS GRILKAANIL IANNPHVFEK RLFSELGYGA ELKVLSANNE EHEAERVTGE LIAHHFVNKT QYKDYAILYR GNHQSRVFEK FLMQNRIPYK ISGGTSFFSR PEIKDLLAYL RVLTNPDDDS AFLRIVNTPK REIGPATLKK LGEWAMTRNK SMFTASFDMG LSQTLSGRGY EALTRFTHWL AEIQRLAERE PIAAVRDLIH GMDYESWLYE TSPSPKAAEM RMKNVNQLFS WMTEMLEGSE LDEPMTLTQV VTRFTLRDMM ERGESEEELD QVQLMTLHAS KGLEFPYVYM VGMEEGFLPH QSSIDEDNID EERRLAYVGI TRAQKELTFT LCKERRQYGE LVRPEPSRFL LELPQDDLIW EQERKVVSAE ERMQKGQSHL ANLKAMMAAK RGK |
| Length |
673 |
| Mol. Wt |
77.026 kDa |
| pI |
7.2 (calculated) |
| Extinction coefficient |
76,780 - 77,405 (calc based on 22 Y, 8 W, and 5 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0012344 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10837 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10837 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000828 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0830 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
Ecoli K-12 |
3.292+/-0.069 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.025729837 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
453 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
102 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
224 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
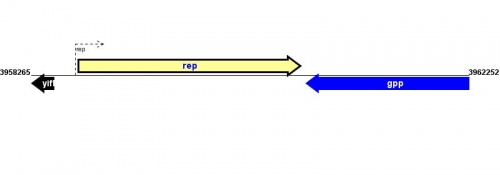 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:3958680..3958720
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for rep | |
|
microarray |
Summary of data for rep from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to rep Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10837 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0830 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10837 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000828 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012344 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Shigella flexneri |
REP |
From SHIGELLACYC |
|
E. coli O157 |
REP |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10837 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10837 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000828 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0830 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012344 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories


