dnaG:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
dnaG |
|---|---|
| Gene Synonym(s) |
ECK3056, b3066, JW3038, dnaP, parB, sdgA[1], sdgA |
| Product Desc. |
Primase for DNA replication; primer synthesis for leading- and lagging-strand synthesis; binds Zn(II)[4] |
| Product Synonyms(s) |
DNA primase[1], B3066[2][1], SdgA[2][1], ParB[2][1], DnaP[2][1], DnaG[2][1] , dnaP, ECK3056, JW3038, parB, sdgA, b3066 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression |
transcription unit(s): rpsU-dnaG-rpoD[2] |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
dnaG |
|---|---|
| Mnemonic |
DNA |
| Synonyms |
ECK3056, b3066, JW3038, dnaP, parB, sdgA[1], sdgA |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
69.17 minutes |
MG1655: 3209129..3210874 |
||
|
NC_012967: 3143924..3145669 |
||||
|
NC_012759: 3096277..3098022 |
||||
|
W3110 |
|
W3110: 3209763..3211508 |
||
|
DH10B: 3306874..3308619 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
3209132 |
Edman degradation |
PMID:6750604 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔdnaG (Keio:JW3038) |
deletion |
deletion |
PMID:16738554 |
||||
|
dnaG::Tn5KAN-I-SceI (FB21017) |
Insertion at nt 798 in Plus orientation |
PMID:15262929 |
does not contain pKD46 | ||||
|
dnaG18 |
|||||||
|
dnaG3(ts) |
temperature sensitive |
PMID:3536848 PMID:3886637 |
|||||
|
dnaG308(ts) |
temperature sensitive |
||||||
|
dnaG399(ts) |
temperature sensitive |
||||||
|
dnaG2903 |
Growth Phenotype |
phage gamma could not multiply at 42C; TS revertants were spontaneously produced at high frequency; at high temperatures, cell growth and DNA replication is halted |
PMID:3886637 |
previously named dnaP18 | |||
| edit table |
<protect></protect>
Notes
The Keio collection[5] lists a deletion of dnaG. The insertion in this strain is a duplication of the dnaG region.[6]
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW3038 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCGCTGGACGAATCCCACGCGT Primer 2:CCtTTTTTCGCCAGCTCCTGGTT | |
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 30% [8] | ||
|
Linked marker |
est. P1 cotransduction: 81% [8] | ||
|
pGNG1 |
Plasmid Clone |
PMID:2055480 | |
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10239 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10239 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000233 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0235 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010065 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
DnaG |
|---|---|
| Synonyms |
DNA primase[1], B3066[2][1], SdgA[2][1], ParB[2][1], DnaP[2][1], DnaG[2][1] , dnaP, ECK3056, JW3038, parB, sdgA, b3066 |
| Product description |
Primase for DNA replication; primer synthesis for leading- and lagging-strand synthesis; binds Zn(II)[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0003676 |
nucleic acid binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006154 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002694 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003896 |
DNA primase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002694 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003896 |
DNA primase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006295 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003896 |
DNA primase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013173 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005658 |
alpha DNA polymerase:primase complex |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0639 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006259 |
DNA metabolic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006154 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006260 |
DNA replication |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002694 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006260 |
DNA replication |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0235 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006269 |
DNA replication, synthesis of RNA primer |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006295 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006269 |
DNA replication, synthesis of RNA primer |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013173 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006269 |
DNA replication, synthesis of RNA primer |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0639 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006350 |
transcription |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0804 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008270 |
zinc ion binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002694 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008270 |
zinc ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0862 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008270 |
zinc ion binding |
PMID:1511009 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0006269 |
DNA replication, synthesis of RNA primer |
PMID:340457 |
IDA: Inferred from Direct Assay |
P |
complete | |||
|
GO:0005737 |
cytoplasm |
PMID:340457 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0003896 |
DNA primase activity |
PMID:340457 |
IDA: Inferred from Direct Assay |
F |
complete | |||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of primosome |
could be indirect |
||
|
Protein |
rplA |
PMID:15690043 |
Experiment(s):EBI-885817 | |
|
Protein |
nusG |
PMID:15690043 |
Experiment(s):EBI-890837 | |
|
Protein |
rplL |
PMID:15690043 |
Experiment(s):EBI-890837 | |
|
Protein |
rpsG |
PMID:15690043 |
Experiment(s):EBI-890837 | |
|
Protein |
yfiF |
PMID:15690043 |
Experiment(s):EBI-890837 | |
|
Protein |
ygdH |
PMID:15690043 |
Experiment(s):EBI-890837 | |
|
Protein |
rho |
PMID:16606699 |
Experiment(s):EBI-1144696 | |
|
Protein |
flu |
PMID:16606699 |
Experiment(s):EBI-1144696 | |
|
Protein |
aspS |
PMID:16606699 |
Experiment(s):EBI-1144696 | |
|
Protein |
ybcW |
PMID:16606699 |
Experiment(s):EBI-1144696 | |
|
Protein |
ygdH |
PMID:19402753 |
LCMS(ID Probability):99.0 | |
|
Protein |
DnaB |
PMID:8917517 |
ELISA | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MAGRIPRVFI NDLLARTDIV DLIDARVKLK KQGKNFHACC PFHNEKTPSF TVNGEKQFYH CFGCGAHGNA IDFLMNYDKL EFVETVEELA AMHNLEVPFE AGSGPSQIER HQRQTLYQLM DGLNTFYQQS LQQPVATSAR QYLEKRGLSH EVIARFAIGF APPGWDNVLK RFGGNPENRQ SLIDAGMLVT NDQGRSYDRF RERVMFPIRD KRGRVIGFGG RVLGNDTPKY LNSPETDIFH KGRQLYGLYE AQQDNAEPNR LLVVEGYMDV VALAQYGINY AVASLGTSTT ADHIQLLFRA TNNVICCYDG DRAGRDAAWR ALETALPYMT DGRQLRFMFL PDGEDPDTLV RKEGKEAFEA RMEQAMPLSA FLFNSLMPQV DLSTPDGRAR LSTLALPLIS QVPGETLRIY LRQELGNKLG ILDDSQLERL MPKAAESGVS RPVPQLKRTT MRILIGLLVQ NPELATLVPP LENLDENKLP GLGLFRELVN TCLSQPGLTT GQLLEHYRGT NNAATLEKLS MWDDIADKNI AEQTFTDSLN HMFDSLLELR QEELIARERT HGLSNEERLE LWTLNQELAK K |
| Length |
581 |
| Mol. Wt |
65.566 kDa |
| pI |
5.9 (calculated) |
| Extinction coefficient |
45,840 - 46,715 (calc based on 16 Y, 4 W, and 7 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
- Purification of DnaG is described in reference[9]
- Km data
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0010065 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10239 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10239 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000233 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0235 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
356 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
98 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
181 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
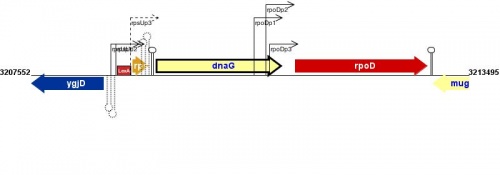 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:3209109..3209149
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for dnaG | |
|
microarray |
Summary of data for dnaG from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to dnaG Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10239 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0235 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10239 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000233 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010065 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Shigella flexneri |
DNAG |
From SHIGELLACYC |
|
E. coli O157 |
DNAG |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10239 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10239 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000233 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0235 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010065 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ Baba, T et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2 2006.0008 PubMed
- ↑ Yamamoto, N et al. (2009) Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 5 335 PubMed
- ↑ 7.0 7.1 CGSC: The Coli Genetics Stock Center
- ↑ 8.0 8.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Rowen, L & Kornberg, A (1978) Primase, the dnaG protein of Escherichia coli. An enzyme which starts DNA chains. J. Biol. Chem. 253 758-64 PubMed
Categories


