cheA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
cheA |
|---|---|
| Gene Synonym(s) |
ECK1889, b1888, JW1877[1], JW1877 |
| Product Desc. |
fused chemotactic sensory histidine kinase in two-component regulatory system with CheB and CheY[1] Histidine protein kinase sensor of chemotactic response; CheY is cognate response regulator; autophosphorylating; CheAS is a short form produced by an internal start at codon 98[2] |
| Product Synonyms(s) |
B1888, ECK1889, JW1877, b1888 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression |
transcription unit(s): motAB-cheAW[3] |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
CheAS is required for polar localization of CheZ. Fla regulon.[2]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
cheA |
|---|---|
| Mnemonic |
Chemotaxis |
| Synonyms |
ECK1889, b1888, JW1877[1], JW1877 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
42.49 minutes |
MG1655: 1973348..1971384 |
||
|
NC_012967: 1954094..1952130 |
||||
|
NC_012759: 1863443..1865407 |
||||
|
W3110 |
|
W3110: 1977038..1975074 |
||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
1971387 |
Edman degradation |
PMID:2002011 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
Strain DH10B |
A34V |
Nonsynonomous mutation |
PMID:18245285 |
This is a SNP in E. coli K-12 Strain DH10B | |||
|
ΔcheA (Keio:JW1877) |
deletion |
deletion |
PMID:16738554 |
||||
|
cheA::Tn5KAN-2 (FB20494) |
Insertion at nt 522 in Minus orientation |
PMID:15262929 |
contains pKD46 | ||||
|
cheA::Tn5KAN-2 (FB20495) |
Insertion at nt 522 in Minus orientation |
PMID:15262929 |
does not contain pKD46 | ||||
|
ΔcheA741::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW1877 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAGCATGGATATAAGCGATTT Primer 2:CCGGCGGCGGTGTTCGCCATACG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 17% [5] | ||
|
Linked marker |
est. P1 cotransduction: 50% [5] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10146 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10146 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000142 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0144 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0006294 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
CheA |
|---|---|
| Synonyms |
B1888, ECK1889, JW1877, b1888 |
| Product description |
fused chemotactic sensory histidine kinase in two-component regulatory system with CheB and CheY[1] Histidine protein kinase sensor of chemotactic response; CheY is cognate response regulator; autophosphorylating; CheAS is a short form produced by an internal start at codon 98[2] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
There are two protein products produced from the cheA gene: CheA(long) and CheA(short).[6]
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0016310 |
phosphorylation |
PMID:2558046 |
IDA: Inferred from Direct Assay |
P |
Figure 3 |
complete | ||
|
GO:0000155 |
two-component sensor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004105 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000155 |
two-component sensor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004105 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000155 |
two-component sensor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005467 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000155 |
two-component sensor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005467 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000160 |
two-component signal transduction system (phosphorelay) |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004105 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000160 |
two-component signal transduction system (phosphorelay) |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004105 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000160 |
two-component signal transduction system (phosphorelay) |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008207 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000160 |
two-component signal transduction system (phosphorelay) |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008207 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000160 |
two-component signal transduction system (phosphorelay) |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0902 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000160 |
two-component signal transduction system (phosphorelay) |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0902 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004673 |
protein histidine kinase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004105 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046777 |
protein amino acid autophosphorylation |
PMID:2558046 |
IDA: Inferred from Direct Assay |
P |
Figure 3 |
complete | ||
|
GO:0004673 |
protein histidine kinase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004105 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004673 |
protein histidine kinase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005467 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004673 |
protein histidine kinase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005467 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000160 |
two-component signal transduction system (phosphorelay) |
PMID:2558046 |
IDA: Inferred from Direct Assay |
P |
with OmpR- Figure 4 |
complete | ||
|
GO:0004673 |
protein histidine kinase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:2.7.13.3 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004673 |
protein histidine kinase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:2.7.13.3 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004871 |
signal transducer activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002545 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004871 |
signal transducer activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002545 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004871 |
signal transducer activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008207 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004871 |
signal transducer activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008207 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004871 |
signal transducer activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR009082 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004871 |
signal transducer activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR009082 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003594 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003594 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005622 |
intracellular |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002545 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005622 |
intracellular |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002545 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004105 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004105 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0963 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0963 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0086 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0086 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006935 |
chemotaxis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002545 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006935 |
chemotaxis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002545 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006935 |
chemotaxis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004105 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006935 |
chemotaxis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004105 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006935 |
chemotaxis |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0145 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006935 |
chemotaxis |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0145 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006935 |
chemotaxis |
PMID:8820640 |
IMP: Inferred from Mutant Phenotype |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002545 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002545 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005467 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005467 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR009082 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR009082 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016301 |
kinase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0418 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016301 |
kinase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0418 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016310 |
phosphorylation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004358 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016310 |
phosphorylation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004358 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016740 |
transferase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0808 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016740 |
transferase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0808 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016772 |
transferase activity, transferring phosphorus-containing groups |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004358 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016772 |
transferase activity, transferring phosphorus-containing groups |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004358 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0018106 |
peptidyl-histidine phosphorylation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005467 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0018106 |
peptidyl-histidine phosphorylation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005467 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0035307 |
positive regulation of protein amino acid dephosphorylation |
PMID:8820640 |
IMP: Inferred from Mutant Phenotype |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0051649 |
establishment of localization in cell |
PMID:12644507 |
IMP: Inferred from Mutant Phenotype |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0051649 |
establishment of localization in cell |
PMID:19502407 |
IDA: Inferred from Direct Assay |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
| edit table |
Interactions
See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of CheA(L) |
could be indirect |
||
|
Protein |
ybaQ |
PMID:16606699 |
Experiment(s):EBI-1141216 | |
|
Protein |
rpmB |
PMID:16606699 |
Experiment(s):EBI-1141216 | |
|
Protein |
nadE |
PMID:16606699 |
Experiment(s):EBI-1141216 | |
|
Protein |
nanE |
PMID:16606699 |
Experiment(s):EBI-1141216 | |
|
Protein |
yfaQ |
PMID:16606699 |
Experiment(s):EBI-1141216 | |
|
Protein |
allC |
PMID:16606699 |
Experiment(s):EBI-1141216 | |
|
Protein |
rplV |
PMID:16606699 |
Experiment(s):EBI-1141216 | |
|
Protein |
appY |
PMID:16606699 |
Experiment(s):EBI-1141216 | |
|
Protein |
fumB |
PMID:16606699 |
Experiment(s):EBI-1141216 | |
|
Protein |
yhbJ |
PMID:16606699 |
Experiment(s):EBI-1141216 | |
|
Protein |
yoaA |
PMID:16606699 |
Experiment(s):EBI-1141216 | |
|
Protein |
Subunits of MCP-II |
could be indirect |
||
|
Protein |
Subunits of MCP-IV |
could be indirect |
||
|
Protein |
Subunits of MCP-III |
could be indirect |
||
|
Protein |
Subunits of MCP-I |
could be indirect |
| |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
cytoplasm |
From EcoCyc[7] |
| ||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MSMDISDFYQ TFFDEADELL ADMEQHLLVL QPEAPDAEQL NAIFRAAHSI KGGAGTFGFS VLQETTHLME NLLDEARRGE MQLNTDIINL FLETKDIMQE QLDAYKQSQE PDAASFDYIC QALRQLALEA KGETPSAVTR LSVVAKSEPQ DEQSRSQSPR RIILSRLKAG EVDLLEEELG HLTTLTDVVK GADSLSAILP GDIAEDDITA VLCFVIEADQ ITFETVEVSP KISTPPVLKL AAEQAPTGRV EREKTTRSNE STSIRVAVEK VDQLINLVGE LVITQSMLAQ RSSELDPVNH GDLITSMGQL QRNARDLQES VMSIRMMPME YVFSRYPRLV RDLAGKLGKQ VELTLVGSST ELDKSLIERI IDPLTHLVRN SLDHGIELPE KRLAAGKNSV GNLILSAEHQ GGNICIEVTD DGAGLNRERI LAKAASQGLT VSENMSDDEV AMLIFAPGFS TAEQVTDVSG RGVGMDVVKR NIQKMGGHVE IQSKQGTGTT IRILLPLTLA ILDGMSVRVA DEVFILPLNA VMESLQPREA DLHPLAGGER VLEVRGEYLP IVELWKVFNV AGAKTEATQG IVVILQSGGR RYALLVDQLI GQHQVVVKNL ESNYRKVPGI SAATILGDGS VALIVDVSAL QAINREQRMA NTAA |
| Length |
654 |
| Mol. Wt |
71.382 kDa |
| pI |
4.7 (calculated) |
| Extinction coefficient |
17,420 - 17,795 (calc based on 8 Y, 1 W, and 3 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0006294 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10146 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10146 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000142 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0144 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
20 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
13 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
0a |
molecules/cell/generation |
|
Ribosome Profiling |
Low confidence in the sequencing data set. |
PMID: 24766808 |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
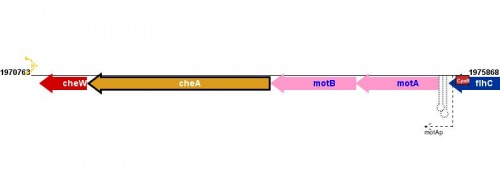 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:1973328..1973368
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for cheA | |
|
microarray |
Summary of data for cheA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to cheA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10146 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0144 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10146 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000142 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0006294 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
CHEA |
From SHIGELLACYC |
|
E. coli O157 |
CHEA |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
PF02895 Signal transducing histidine kinase, homodimeric domain |
||
|
PF02518 Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase |
||
|
EcoCyc:EG10146 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10146 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000142 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0144 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0006294 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 CGSC: The Coli Genetics Stock Center
- ↑ 5.0 5.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Smith, RA & Parkinson, JS (1980) Overlapping genes at the cheA locus of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 77 5370-4 PubMed
- ↑ EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
Categories


