xerC:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
xerC |
|---|---|
| Gene Synonym(s) |
ECK3806, b3811, JW3784[1], JW3784 |
| Product Desc. |
site-specific recombinase, acts on cer sequence of ColE1, effects chromosome segregation at cell division[2][3]; Component of Xer site-specific recombination system[3] Tyrosine recombinase XerCD resolves chromosome dimers; site-specific[4] |
| Product Synonyms(s) |
site-specific tyrosine recombinase[1], B3811[2][1], XerC[2][1] , ECK3806, JW3784, b3811 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
XerCD acts at the Dif site.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
xerC |
|---|---|
| Mnemonic |
cer-specific recombination |
| Synonyms |
ECK3806, b3811, JW3784[1], JW3784 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
86.09 minutes, 86.09 minutes |
MG1655: 3994310..3995206 |
||
|
NC_012967: 3958124..3959020 |
||||
|
NC_012759: 3883979..3884875 |
||||
|
W3110 |
|
W3110: 3640394..3639498 |
||
|
DH10B: 4093230..4094126 |
||||
| edit table |
</protect>
Notes
The xerC gene is part of a multicistronic unit, described in Colloms et al. (1990)[5].
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
3994310 |
Edman degradation |
PMID:2254268 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔxerC (Keio:JW3784) |
deletion |
deletion |
PMID:16738554 |
||||
|
xerCR243Q |
R243Q |
Abolishes DNA cleavage activity |
|
seeded from UniProt:P0A8P6 | |||
|
xerCY275F |
Y275F |
Abolishes DNA cleavage activity |
|
seeded from UniProt:P0A8P6 | |||
|
xerCESS252NHG |
ESS252NHG |
Abolishes chromosomal recombination but not plasmid resolution |
seeded from UniProt:P0A8P6 | ||||
|
xerC2 |
Growth Phenotype |
small colonies on broth-containing plates, slow recovery from stationary phase. Also, "xerC cells in exponential phase show a substantial, but variable # of filaments and an excess of nonfilamentous cells at the 2- cell stage." |
PMID:1931824 |
Exponential growth rate in liquid broth culture showed day-to-day variation that varied from 80-100% of an otherwise isogenic xerC+ strain. | |||
|
xerCY17 |
Growth Phenotype |
small colonies on broth-containing plates, slow recovery from stationary phase. Also, "xerC cells in exponential phase show a substantial, but variable # of filaments and an excess of nonfilamentous cells at the 2- cell stage." |
PMID:1931824 |
Exponential growth rate in liquid broth culture showed day-to-day variation that varied from 80-100% of an otherwise isogenic xerC+ strain. | |||
|
xerC |
Cell Shape |
Filamentation- >15% of cells |
PMID:15065657 |
||||
|
xerC rarA |
Cell Shape |
Filamentation- >15% of cells |
PMID:15065657 |
||||
|
xerC1 |
resolution defective |
PMID:2254268 |
| ||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW3784 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCACCGATTTACACACCGATGT Primer 2:CCTTTCCCCCGTTTGGCGCGTGG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 17% [7] | ||
|
metEo-3079::Tn10 |
Linked marker |
est. P1 cotransduction: 60% [7] | |
|
DS8029 |
Strain |
AB1157xerC |
PMID:10523315 |
|
GR51 |
Strain |
AB1157xerC ftsK |
PMID:10523315 |
|
DW1 |
Strain |
AB1157recF lacIq lacPO-xerC recA938 |
PMID:10523315 |
|
RM40 |
Strain |
AB1157recF lacIq lacPO-xerC |
PMID:8168483 |
|
pSDC107 |
Plasmid Clone |
XerC+ overexpressing plasmid CmR λdv replicon |
PMID:1931824 |
|
pSDC105 |
Plasmid Clone |
XerC+ overexpressing plasmid CmR p15a replicon |
PMID:2254268 |
|
pLA125 |
Plasmid Clone |
XerCR243Q overexpressing plasmid |
PMID:8402918 |
|
pSDC104 |
Plasmid Clone |
XerC+ overexpressing plasmid |
PMID:2254268 |
|
pLA121 |
Plasmid Clone |
XerCY275F overexpressing plasmid |
PMID:8402918 |
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11069 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11069 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001058 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB1062 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012448 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
XerC |
|---|---|
| Synonyms |
site-specific tyrosine recombinase[1], B3811[2][1], XerC[2][1] , ECK3806, JW3784, b3811 |
| Product description |
site-specific recombinase, acts on cer sequence of ColE1, effects chromosome segregation at cell division[2][3]; Component of Xer site-specific recombination system[3] Tyrosine recombinase XerCD resolves chromosome dimers; site-specific[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
XerC binds to the dif left half site[8].
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002104 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0042150 |
plasmid recombination |
PMID:2254268 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004107 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009009 |
site-specific recombinase activity |
PMID:2254268 |
IMP: Inferred from Mutant Phenotype |
F |
complete | |||
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011010 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011931 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013762 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0238 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_01808 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0963 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0086 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006313 |
transposition, DNA-mediated |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_01808 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007049 |
cell cycle |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011931 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007049 |
cell cycle |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0131 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007059 |
chromosome segregation |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_01808 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007059 |
chromosome segregation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011931 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007059 |
chromosome segregation |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0159 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009037 |
tyrosine-based site-specific recombinase activity |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_01808 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0015074 |
DNA integration |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002104 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0015074 |
DNA integration |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004107 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0015074 |
DNA integration |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011931 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0015074 |
DNA integration |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013762 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0015074 |
DNA integration |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0229 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0051301 |
cell division |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011931 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0051301 |
cell division |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0132 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009009 |
site-specific recombinase activity |
PMID:11114887 |
IDA: Inferred from Direct Assay |
F |
with XerD and FtsK |
complete | ||
|
GO:0048476 |
Holliday junction resolvase complex |
PMID:11114887 |
IDA: Inferred from Direct Assay |
C |
with XerD |
complete | ||
|
GO:0006276 |
plasmid maintenance |
PMID:2254268 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0003677 |
DNA binding |
PMID:8402918 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0009037 |
tyrosine-based site-specific recombinase activity |
PMID:8402918 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0006276 |
plasmid maintenance |
PMID:8195072 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0071139 |
resolution of recombination intermediates |
PMID:7746851 |
IDA: Inferred from Direct Assay |
P |
complete | |||
|
GO:0048476 |
Holliday junction resolvase complex |
PMID:7746851 |
IDA: Inferred from Direct Assay |
C |
with XerD |
complete | ||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of Xer site-specific recombination system |
could be indirect |
||
|
Protein |
pldA |
PMID:16606699 |
Experiment(s):EBI-1146722 | |
|
Protein |
cysD |
PMID:16606699 |
Experiment(s):EBI-1146722 | |
|
Protein |
prs |
PMID:16606699 |
Experiment(s):EBI-1146722 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MTDLHTDVER YLRYLSVERQ LSPITLLNYQ RQLEAIINFA SENGLQSWQQ CDVTMVRNFA VRSRRKGLGA ASLALRLSAL RSFFDWLVSQ NELKANPAKG VSAPKAPRHL PKNIDVDDMN RLLDIDINDP LAVRDRAMLE VMYGAGLRLS ELVGLDIKHL DLESGEVWVM GKGSKERRLP IGRNAVAWIE HWLDLRDLFG SEDDALFLSK LGKRISARNV QKRFAEWGIK QGLNNHVHPH KLRHSFATHM LESSGDLRGV QELLGHANLS TTQIYTHLDF QHLASVYDAA HPRAKRGK |
| Length |
298 |
| Mol. Wt |
33.868 kDa |
| pI |
9.8 (calculated) |
| Extinction coefficient |
41,940 - 42,065 (calc based on 6 Y, 6 W, and 1 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Mutants that are still able to bind recombination sites, but do not promote recombination at cer or in vivo plasmid resolution
- Presumptive active site tyrosine nucleophile mutant = XerC275F[9][10]
- Conserved Domain II Arginine implicated in DNA phosphodiester activation = XerCR243Q[11][10]
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0012448 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG11069 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11069 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120001058 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1062 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
393 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
212 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
361 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
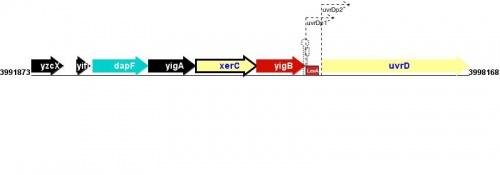 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:3994290..3994330
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for xerC | |
|
microarray |
Summary of data for xerC from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to xerC Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11069 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1062 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11069 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001058 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012448 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
XERC |
From SHIGELLACYC |
|
E. coli O157 |
XERC |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Neilson et al. (1999) describes site-specific recombination at dif by H. influenzae XerC[12]. They conclude the HinXerC and HinXerD can interact with their E.coli counterparts.
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11069 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11069 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001058 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1062 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012448 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Member of the lambda integrase family of site-specific recombinases[5].
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 Colloms, SD et al. (1990) Recombination at ColE1 cer requires the Escherichia coli xerC gene product, a member of the lambda integrase family of site-specific recombinases. J. Bacteriol. 172 6973-80 PubMed
- ↑ 6.0 6.1 CGSC: The Coli Genetics Stock Center
- ↑ 7.0 7.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Blakely, G et al. (1993) Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell 75 351-61 PubMed
- ↑ Pargellis, CA et al. (1988) Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J. Biol. Chem. 263 7678-85 PubMed
- ↑ 10.0 10.1 Leslie, NR & Sherratt, DJ (1995) Site-specific recombination in the replication terminus region of Escherichia coli: functional replacement of dif. EMBO J. 14 1561-70 PubMed
- ↑ Parsons, RL et al. (1988) Step-arrest mutants of FLP recombinase: implications for the catalytic mechanism of DNA recombination. Mol. Cell. Biol. 8 3303-10 PubMed
- ↑ Neilson, L et al. (1999) Site-specific recombination at dif by Haemophilus influenzae XerC. Mol. Microbiol. 31 915-26 PubMed
Categories


