tap:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
tap |
|---|---|
| Gene Synonym(s) |
ECK1886, b1885, JW1874[1], JW1874 |
| Product Desc. |
Dipeptide chemoreceptor, methyl-accepting; MCP IV; flagellar regulon[4] |
| Product Synonyms(s) |
methyl-accepting protein IV[1], B1885[2][1], Tap[2][1] , ECK1886, JW1874, b1885 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression |
transcription unit(s): tar-tap-cheRBYZ[2] |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Tap sense dipeptides indirectly through the periplasmic dipeptide binding protein DppA. Tap is a low-abundance chemoreceptor, 5- 10-fold lower than Tar and Tsr. Tap can function as a cold receptor during thermotaxis. Tap senses low pH as a repellent.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
tap |
|---|---|
| Mnemonic |
Taxis protein |
| Synonyms |
ECK1886, b1885, JW1874[1], JW1874 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
42.4 minutes |
MG1655: 1969008..1967407 |
||
|
NC_012967: 1949754..1948153 |
||||
|
NC_012759: 1859466..1861067 |
||||
|
W3110 |
|
W3110: 1972698..1971097 |
||
|
DH10B: 2059579..2057978 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
Δtap (Keio:JW1874) |
deletion |
deletion |
PMID:16738554 |
||||
|
tap::Tn5KAN-2 (FB20489) |
Insertion at nt 194 in Minus orientation |
PMID:15262929 |
does not contain pKD46 | ||||
|
tap::Tn5KAN-2 (FB20490) |
Insertion at nt 194 in Minus orientation |
PMID:15262929 |
contains pKD46 | ||||
|
Δtap::kan |
deletion |
Biolog:respiration |
unable to respire Acetate |
PMID:16095938 |
|||
|
Δtap::kan |
deletion |
Biolog:respiration |
unable to respire a-Ketoglutarate |
PMID:16095938 |
|||
|
Δtap::kan |
deletion |
Biolog:respiration |
unable to respire L-Aspartate |
PMID:16095938 |
|||
|
Δtap-738::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW1874 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCTTTAATCGTATTCGAATTTC Primer 2:CCGGATACCACTGGCGCAATTTG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 21% [6] | ||
|
Linked marker |
est. P1 cotransduction: 43% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10987 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10987 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000976 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0980 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0006288 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
Tap |
|---|---|
| Synonyms |
methyl-accepting protein IV[1], B1885[2][1], Tap[2][1] , ECK1886, JW1874, b1885 |
| Product description |
Dipeptide chemoreceptor, methyl-accepting; MCP IV; flagellar regulon[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0004871 |
signal transducer activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003660 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004871 |
signal transducer activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004089 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004871 |
signal transducer activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004090 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004871 |
signal transducer activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004091 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004871 |
signal transducer activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0807 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004888 |
transmembrane receptor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003122 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0997 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-1003 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006935 |
chemotaxis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003122 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006935 |
chemotaxis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004089 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006935 |
chemotaxis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004090 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006935 |
chemotaxis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004091 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006935 |
chemotaxis |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0145 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003122 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003660 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004089 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004090 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004091 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0807 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003122 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004089 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004090 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004091 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0472 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003660 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0812 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-9909 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of MCP-IV |
could be indirect |
| |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
plasma membrane |
C-terminus localized in the cytoplasm with 2 predicted transmembrane domains |
Daley et al. (2005) [7] |
||
|
plasma membrane |
From EcoCyc[3] |
| ||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MFNRIRISTT LFLILILCGI LQIGSNGMSF WAFRDDLQRL NQVEQSNQQR AALAQTRAVM LQASTALNKA GTLTALSYPA DDIKTLMTTA RASLTQSTTL FKSFMAMTAG NEHVRGLQKE TEKSFARWHN DLEHQATWLE SNQLSDFLTA PVQGSQNAFD VNFEAWQLEI NHVLEAASAQ SQRNYQISAL VFISMIIVAA IYISSALWWT RKMIVQPLAI IGSHFDSIAA GNLARPIAVY GRNEITAIFA SLKTMQQALR GTVSDVRKGS QEMHIGIAEI VAGNNDLSSR TEQQAASLAQ TAASMEQLTA TVGQNADNAR QASELAKNAA TTAQAGGVQV STMTHTMQEI ATSSQKIGDI ISVIDGIAFQ TNILALNAAV EAARAGEQGR GFAVVAGEVR NLASRSAQAA KEIKGLIEES VNRVQQGSKL VNNAAATMID IVSSVTRVND IMGEIASASE EQQRGIEQVA QAVSQMDQVT QQNASLVEEA AVATEQLANQ ADHLSSRVAV FTLEEHEVAR HESVQLQIAP VVS |
| Length |
533 |
| Mol. Wt |
57.511 kDa |
| pI |
5.6 (calculated) |
| Extinction coefficient |
38,960 - 39,085 (calc based on 4 Y, 6 W, and 1 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0006288 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10987 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10987 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000976 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0980 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
0a |
molecules/cell/generation |
|
Ribosome Profiling |
Low confidence in the sequencing data set. |
PMID: 24766808 |
|
Protein |
E. coli K-12 MG1655 |
2a |
molecules/cell/generation |
|
Ribosome Profiling |
Low confidence in the sequencing data set. |
PMID: 24766808 |
|
Protein |
E. coli K-12 MG1655 |
0a |
molecules/cell/generation |
|
Ribosome Profiling |
Low confidence in the sequencing data set. |
PMID: 24766808 |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
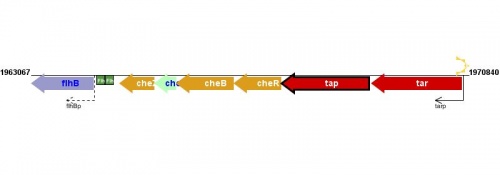 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:1968988..1969028
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for tap | |
|
microarray |
Summary of data for tap from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to tap Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10987 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0980 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10987 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000976 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0006288 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
TAP |
From SHIGELLACYC |
|
E. coli O157 |
TAP |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
PF00015 Methyl-accepting chemotaxis protein (MCP) signaling domain |
||
|
EcoCyc:EG10987 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10987 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000976 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0980 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0006288 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 3.4 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Daley, DO et al. (2005) Global topology analysis of the Escherichia coli inner membrane proteome. Science 308 1321-3 PubMed
Categories



