recA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
recA |
|---|---|
| Gene Synonym(s) |
ECK2694, b2699, JW2669, lexB, srf, umuB, zab, recH, rnmB, tif, umuR[1], umuR |
| Product Desc. |
DNA strand exchange and renaturation, DNA-dependent ATPase, DNA- and ATP-dependent coprotease[2]; DNA strand exchange and recombination protein with protease and nuclease activity[3] General recombination and DNA repair; pairing and strand exchange; role in cleavage of LexA repressor, SOS mutagenesis[4] |
| Product Synonyms(s) |
B2699[2][1], Tif[2][1], UmuB[2][1], Zab[2][1], Srf[2][1], RnmB[2][1], RecH[2][1], LexB[2][1], RecA[2][1] , ECK2694, JW2669, lexB, recH, rnmB, srf, tif, umuB, umuR, zab, b2699 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
RecA does not bind to DNA with extensive stem-loop structures, except in the presence of SSB. RecA is required for polV lesion bypass. RecA filamentation is inhibited by RecX. Binds TrxA (Kumar, 2004).[4]
RecA lacking mutants exhibited the same frequency as the wild type when exposed to sublethal concentrations of the antibiotics Ampicillin, Ceftazidime, Imipenem, Fosfomycin, Ciprofloxacin, Trimethoprim, and Sulfamethoxazole.(PMID:21212055)
RecA lacking mutants exhibited different frequencies then the wild type when exposed to sublethal concentrations of the antibiotics Gentamicin, Rifampicin, Chloramphenicol (PMID:21212055)
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
recA |
|---|---|
| Mnemonic |
Recombination |
| Synonyms |
ECK2694, b2699, JW2669, lexB, srf, umuB, zab, recH, rnmB, tif, umuR[1], umuR |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
60.8 minutes |
MG1655: 2821791..2820730 |
||
|
NC_012967: 2718628..2717567 |
||||
|
NC_012759: 2706542..2707603 |
||||
|
W3110 |
|
W3110: 2822425..2821364 |
||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
2820733 |
Edman degradation |
PMID:6244554 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
Strain DH10B |
G161D |
Nonsynonomous mutation |
PMID:18245285 |
This is a SNP in E. coli K-12 Strain DH10B | |||
|
recA(del) (Keio:JW2669) |
deletion |
deletion |
PMID:16738554 |
||||
|
recA::Tn5KAN-I-SceI (FB20884) |
Insertion at nt 407 in Plus orientation |
PMID:15262929 |
does not contain pKD46 | ||||
|
recA::Tn5KAN-I-SceI (FB20885) |
Insertion at nt 407 in Plus orientation |
PMID:15262929 |
contains pKD46 | ||||
|
recA(del)::kan |
deletion-replacement |
Biolog:respiration |
unable to respire a-Ketoglutarate |
PMID:16095938 |
|||
|
recA(del)::kan |
deletion |
Biolog:respiration |
unable to respire Succinate |
PMID:16095938 |
|||
|
recA(del)::kan |
deletion |
Biolog:respiration |
unable to respire L-Asparagine |
PMID:16095938 |
|||
|
recA938::Tn9-200 |
|||||||
|
recA1 |
PMID:6363880 PMID:350827 |
||||||
|
recA56 |
PMID:2651400 |
||||||
|
recA13 |
PMID:2651400 |
||||||
|
recA44(ts) |
temperature sensitive |
PMID:2651400 |
|||||
|
recA41 |
|||||||
|
recA441(ts) |
temperature sensitive |
PMID:6363880 PMID:2406267 PMID:350827 |
|||||
|
recA430 |
PMID:6363880 PMID:2651400 PMID:1400177 |
||||||
|
recA3 |
|||||||
|
recA11 |
PMID:7512687 PMID:5321955 PMID:350827 |
||||||
|
recA200(ts) |
temperature sensitive |
||||||
|
recA14 |
|||||||
|
recA12 |
PMID:7512687 PMID:350827 |
||||||
|
recA431 |
|||||||
|
recA34 |
|||||||
|
recA99(Am) |
PMID:350827 PMID:2651400 |
amber (UAG) mutation | |||||
|
recA142 |
PMID:2651400 |
||||||
|
recA111 |
|||||||
|
recA629 |
PMID:6088537 |
||||||
|
recA2111::RP4-2Tc::Mu |
|||||||
|
recA635(del)::kan |
PMID:10829079 |
||||||
|
recA636(del-ins)::FRT |
PMID:10829079 |
||||||
|
recA2 |
|||||||
|
recA1907::cat-aadA |
PMID:2155195 |
||||||
|
recA774(del)::kan |
PMID:16738554 |
||||||
|
recA730 |
constitutively activated form of RecA so it cleaves the LexA repressor in the absence of SOS induction |
PMID:2834329 |
|||||
|
recA |
deletion |
Sensitivity to |
increased sensitivity to Bicyclomycin |
PMID:21357484 |
fig 2 | ||
|
recA(del) |
deletion |
Growth Phenotype |
When the recA gene was deleted there was only a marginal change in the growth rate compared to the wild type when exposed to the same concentration of levofloxacin. |
PMID:20660686 |
See figure 1. | ||
|
recA(del) |
The recA was deleted. |
Resistant to |
After the recA gene was deleted there was only a marginal difference in the level of resistance compared to the wild type strain when exposed to the same concentration of levofloxacin. |
PMID:20660686 |
Figure 1B. | ||
|
recA(del) |
The recA gene was deleted. |
Resistant to |
The number of recA(del) levofloxacin resistant mutants was fewer than the wildtype and did not appear as quickly. |
PMID:20660686 |
See Figure 4. | ||
|
recA(del) |
The recA gene was deactivated. |
Sensitivity to |
Mutant frequency decreased with the introduction of sublethal dosages of antibiotics. |
PMID:21212055 |
See Figure 1 | ||
|
recA(del) |
The recA gene was deactivated. |
Rate transcription |
the rate of transcription of recA::GFP was increased with the additon antimicrobials |
PMID:21212055 |
See figure 2 | ||
|
recA(del) |
The recA gene was deactivated. |
Growth Phenotype |
The addition of the antimicrobials ciprofloxacin, trimethoprim and sulfamethoxazole to the recA deficient mutant caused the formation of reduced filaments. |
PMID:21212055 |
See figure 3(b) | ||
|
nrdA101recA(del) |
'recA is deactivated. |
rate of replication |
Replication is stalled when the replication forks are reversed due to the absence of recA |
PMID:21441507 |
|||
|
recA(del) |
the recA gene was deactiaved. |
Sensitivity to |
By deactivating the recA gene there is an increase in sensitvity to UV radiation. |
PMID:2112525 |
See Figure 2 A. | ||
|
recA(del) |
the recA gene was deactiaved. |
Rate of Recombination |
By deactivating the recA gene there is a decrease in the rate of recombination frequency compared to the wildtype strain. |
PMID:2112525 |
See Figure 2 B. | ||
|
recA(del) |
the recA gene was deactiaved. |
rate of LexA cleavage |
By deactivating the recA gene there is a decrease LexA cleavage when compared to the wildtype. |
PMID:2112525 |
See Figure 2 C. | ||
|
recA142 |
Sensitivity to |
mutants are more sensitive to DNA damaging agent Mitomycin C, fig 2. |
PMID:17040104 |
||||
|
redA in strain F19 |
Resistant to |
Resistant to Muteonidazole resistance |
PMID:1991710 |
Strain: F19 |
Figure 1 | ||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW2669 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCGCTATCGACGAAAACAAACA Primer 2:CCAAAATCTTCGTTAGTTTCTGC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: % [6] | ||
|
Linked marker |
est. P1 cotransduction: 85% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10823 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10823 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000814 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0816 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0008876 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
RecA |
|---|---|
| Synonyms |
B2699[2][1], Tif[2][1], UmuB[2][1], Zab[2][1], Srf[2][1], RnmB[2][1], RecH[2][1], LexB[2][1], RecA[2][1] , ECK2694, JW2669, lexB, recH, rnmB, srf, tif, umuB, umuR, zab, b2699 |
| Product description |
DNA strand exchange and renaturation, DNA-dependent ATPase, DNA- and ATP-dependent coprotease[2]; DNA strand exchange and recombination protein with protease and nuclease activity[3] General recombination and DNA repair; pairing and strand exchange; role in cleavage of LexA repressor, SOS mutagenesis[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0003697 |
single-stranded DNA binding |
PMID:19820696 |
IDA: Inferred from Direct Assay |
F |
CACAO annotation. Earlier reference would be preferable |
complete | ||
|
GO:0000166 |
nucleotide binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003593 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001553 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020584 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020587 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020588 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0238 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003684 |
damaged DNA binding |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00268 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003697 |
single-stranded DNA binding |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00268 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003697 |
single-stranded DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013765 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00268 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001553 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013765 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020584 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020587 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020588 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00268 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013765 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0048870 |
cell motility |
PMID:17391508 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0005737 |
cytoplasm |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0963 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0086 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006259 |
DNA metabolic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001553 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006259 |
DNA metabolic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020584 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006259 |
DNA metabolic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020587 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006259 |
DNA metabolic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020588 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006281 |
DNA repair |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00268 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006281 |
DNA repair |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013765 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006974 |
response to DNA damage stimulus |
PMID:10760155 |
IEP: Inferred from Expression Pattern |
P |
complete | |||
|
GO:0006281 |
DNA repair |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0234 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006310 |
DNA recombination |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00268 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006310 |
DNA recombination |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0233 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006950 |
response to stress |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0346 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006974 |
response to DNA damage stimulus |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00268 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006974 |
response to DNA damage stimulus |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0227 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008094 |
DNA-dependent ATPase activity |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00268 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008094 |
DNA-dependent ATPase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001553 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008094 |
DNA-dependent ATPase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020584 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008094 |
DNA-dependent ATPase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020587 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008094 |
DNA-dependent ATPase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020588 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009432 |
SOS response |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00268 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009432 |
SOS response |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0742 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0017111 |
nucleoside-triphosphatase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003593 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006974 |
response to DNA damage stimulus |
PMID:11967071 |
IEP: Inferred from Expression Pattern |
P |
complete | |||
|
GO:0009432 |
SOS response |
PMID:4600265 |
IMP: Inferred from Mutant Phenotype |
P |
Fig. 1 and 2 |
complete | ||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
napC |
PMID:16606699 |
Experiment(s):EBI-1143694 | |
|
Protein |
rplJ |
PMID:16606699 |
Experiment(s):EBI-1143694 | |
|
Protein |
nadE |
PMID:16606699 |
Experiment(s):EBI-1143694 | |
|
Protein |
rplB |
PMID:16606699 |
Experiment(s):EBI-1143694 | |
|
Protein |
yfiR |
PMID:16606699 |
Experiment(s):EBI-1143694 | |
|
Protein |
groS |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
fusA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MAIDENKQKA LAAALGQIEK QFGKGSIMRL GEDRSMDVET ISTGSLSLDI ALGAGGLPMG RIVEIYGPES SGKTTLTLQV IAAAQREGKT CAFIDAEHAL DPIYARKLGV DIDNLLCSQP DTGEQALEIC DALARSGAVD VIVVDSVAAL TPKAEIEGEI GDSHMGLAAR MMSQAMRKLA GNLKQSNTLL IFINQIRMKI GVMFGNPETT TGGNALKFYA SVRLDIRRIG AVKEGENVVG SETRVKVVKN KIAAPFKQAE FQILYGEGIN FYGELVDLGV KEKLIEKAGA WYSYKGEKIG QGKANATAWL KDNPETAKEI EKKVRELLLS NPNSTPDFSV DDSEGVAETN EDF |
| Length |
353 |
| Mol. Wt |
37.974 kDa |
| pI |
5.0 (calculated) |
| Extinction coefficient |
21,430 - 21,805 (calc based on 7 Y, 2 W, and 3 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0008876 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10823 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10823 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000814 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0816 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
6.86E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
E. coli K-12 MG1655 |
10377 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
2926 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
5850 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
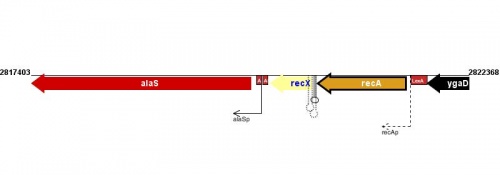 Figure courtesy of RegulonDB |
</protect>
Notes
LexA-regulated[7]
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:2821771..2821811
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for recA | |
|
microarray |
Summary of data for recA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (2821707..2821893) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to recA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10823 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0816 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10823 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000814 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0008876 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Saccharomyces cerevisiae |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
RECA |
From SHIGELLACYC |
|
E. coli O157 |
RECA |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10823 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10823 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000814 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0816 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0008876 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 2.18 2.19 2.20 2.21 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Fernández De Henestrosa, AR et al. (2000) Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35 1560-72 PubMed
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories
- Genes in OpenBioSystems with Promoter Fusions
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Canis familiaris
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Homo sapiens
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Oryza gramene
- Genes with homologs in Pan troglodytes
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Saccharomyces cerevisiae
- Genes with homologs in Schizosaccharomyces pombe
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


