nuoG:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
nuoG |
|---|---|
| Gene Synonym(s) |
ECK2277, b2283, JW2278[1], JW2278 |
| Product Desc. |
Component of soluble NADH dehydrogenase fragment[3]; NADH dehydrogenase I[2][3] NADH:ubiquinone oxidoreductase subunit G, complex I; NADH dehydrogenase I[4] |
| Product Synonyms(s) |
NADH:ubiquinone oxidoreductase, chain G[1], B2283[2][1], NuoG[2][1] , ECK2277, JW2278, b2283 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression |
transcription unit(s): nuoABCEFGHIJKLMN[2], nuo1-14 |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Leif et al. (95) report that the initial Met is not clipped, in conflict with Link et al. (97), who found it was clipped. M-A is almost always clipped, so Link et al. seems to be the more reliable data.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
nuoG |
|---|---|
| Mnemonic |
NADH:ubiquinone oxidoreductase |
| Synonyms |
ECK2277, b2283, JW2278[1], JW2278 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
51.63 minutes |
MG1655: 2398193..2395461 |
||
|
NC_012967: 2343762..2341030 |
||||
|
NC_012759: 2281266..2283992 |
||||
|
W3110 |
|
W3110: 2404841..2402109 |
||
|
DH10B: 2489958..2487226 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
2395464 |
Edman degradation |
PMID:9298646 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
nuoG(del) (Keio:JW2278) |
deletion |
deletion |
PMID:16738554 |
||||
|
nuoG::Tn5KAN-2 (FB20742) |
Insertion at nt 519 in Minus orientation |
PMID:15262929 |
does not contain pKD46 | ||||
|
nuoG::Tn5KAN-2 (FB20743) |
Insertion at nt 519 in Minus orientation |
PMID:15262929 |
contains pKD46 | ||||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire D-Galactose |
PMID:16095938 |
|||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire b-Methyl-D-glucoside |
PMID:16095938 |
|||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire Acetate |
PMID:16095938 |
|||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire a-Hydroxybutyrate |
PMID:16095938 |
|||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire a-Ketobutyrate |
PMID:16095938 |
|||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire a-Ketoglutarate |
PMID:16095938 |
|||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire Succinate |
PMID:16095938 |
|||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire Bromosuccinate |
PMID:16095938 |
|||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire Glucuronamide |
PMID:16095938 |
|||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire L-Alanine |
PMID:16095938 |
|||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire L-Alanyl-glycine |
PMID:16095938 |
|||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire L-Asparagine |
PMID:16095938 |
|||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire L-Aspartate |
PMID:16095938 |
|||
|
nuoG(del)::kan |
deletion |
Biolog:respiration |
unable to respire L-Serine |
PMID:16095938 |
|||
|
nuoG765(del)::kan |
PMID:16738554 |
||||||
|
nuoG::Tn10dCam |
Insertion |
Mutagenesis Rate |
Decrease in stress induced mutagenesis (SIM). |
PMID:23224554 |
Parent Strain: SMR4562 Experimental Strain: SMR13640 |
The Mutation conferred a strong decrease in SIM with mutant frequency being decreased by over 90 percent. See Table S3 for full experimental data. | |
|
nuoG::Tn10dCam |
Insertion |
SDS-EDTA Sensitivity |
PMID:23224554 |
Parent Strain: SMR4562 Experimental Strain: SMR13640 |
The Mutation conferred an increase in SDS-EDTA sensitivity. See table S7 and S1 for a summary of experimental results. | ||
|
SMR4562 yiaG-yfp FRT nuoG::Tn10dCam |
insertion |
SigmaS activity |
Decrease in sigmaS activity |
PMID:23224554 |
Parental Strain: SMR10582 Experimental Strain: SMR14420 |
See table S8 for full experimental results. | |
|
CAG45114 nuoG::Tn10dCam |
Insertion |
SigmaE activity |
Decrease in SigmaE activity |
PMID:23224554 |
Parental Strain: CAG45114 Experimental Strain: SMR16038 |
See table S11 for experimental results. | |
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW2278 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCCTAATGGCTACAATTCATGT Primer 2:CCTTGTTGTGCCTCCTTGAGATC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 82% [6] | ||
|
zfd-1::Tn10 |
Linked marker |
est. P1 cotransduction: % [6] | |
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG12087 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12087 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001992 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB2011 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0007543 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
NuoG |
|---|---|
| Synonyms |
NADH:ubiquinone oxidoreductase, chain G[1], B2283[2][1], NuoG[2][1] , ECK2277, JW2278, b2283 |
| Product description |
Component of soluble NADH dehydrogenase fragment[3]; NADH dehydrogenase I[2][3] NADH:ubiquinone oxidoreductase subunit G, complex I; NADH dehydrogenase I[4] |
| EC number (for enzymes) |
1.6.5.3[1] |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
NOT |
GO:0005737 |
cytoplasm |
PMID:7607227 |
IDA: Inferred from Direct Assay |
C |
Seeded from Riley et al 2006 [1]. See discussion page for explanation of 'NOT' qualifier. |
complete | |
|
GO:0030964 |
NADH dehydrogenase complex |
PMID:7607227 |
IDA: Inferred from Direct Assay |
C |
The E. coli NADH dehydrogenase complex was purified and the subunits separated by SDS-PAGE. The N-termini of these 6 polypeptides were sequenced after deblocking with methanolic HCl. |
complete | ||
|
GO:0005886 |
plasma membrane |
PMID:7607227 |
IDA: Inferred from Direct Assay |
C |
The NADH dehydrogenase I complex fractionates with cytoplasmic membranes. |
complete | ||
|
GO:0005506 |
iron ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0408 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
Contributes to |
GO:0003954 |
NADH dehydrogenase activity |
PMID:3122832 |
IDA: Inferred from Direct Assay |
F |
Purified NADH dehydrogenase I in membrane vesicles can oxidize NADH. |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0963 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0051539 |
4 iron, 4 sulfur cluster binding |
PMID:17640900 |
IDA: Inferred from Direct Assay |
F |
EPR spectra from purified NuoG reduced by dithionite gave evidence of two rhombic [4Fe-4S] clusters (Fig. 2 and text). |
complete | ||
|
GO:0005737 |
cytoplasm |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0086 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0051537 |
2 iron, 2 sulfur cluster binding |
PMID:17640900 |
IDA: Inferred from Direct Assay |
F |
EPR spectra from purified NuoG reduced by dithionite gave evidence of an axial [2Fe-2S] cluster (Fig. 2 and text). |
complete | ||
|
GO:0008137 |
NADH dehydrogenase (ubiquinone) activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000283 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
Contributes to |
GO:0010181 |
FMN binding |
PMID:7607227 |
IDA: Inferred from Direct Assay |
F |
The water-soluble NADH fragment contains the subunits NuoE, NuoF, and NuoG. This fragment also contains FMN and has the EPR-detectable FeS clusters N1b, N1c, N3, and N4. |
complete | |
|
GO:0009055 |
electron carrier activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001041 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016491 |
oxidoreductase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006656 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016491 |
oxidoreductase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006657 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016491 |
oxidoreductase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006963 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016491 |
oxidoreductase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0560 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016651 |
oxidoreductase activity, acting on NADH or NADPH |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR010228 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0019898 |
extrinsic to membrane |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-9903 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030151 |
molybdenum ion binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006657 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0042773 |
ATP synthesis coupled electron transport |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000283 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046872 |
metal ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0479 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0048038 |
quinone binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0874 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0050136 |
NADH dehydrogenase (quinone) activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:1.6.99.5 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0051537 |
2 iron, 2 sulfur cluster binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0001 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0051539 |
4 iron, 4 sulfur cluster binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0004 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000283 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR010228 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0560 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of soluble NADH dehydrogenase fragment |
could be indirect |
||
|
Protein |
skp |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
leuD |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
nadC |
PMID:19402753 |
LCMS(ID Probability):99.3 | |
|
Protein |
purA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
purL |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
nuoF |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
nuoE |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
Subunits of NADH dehydrogenase I |
could be indirect |
| |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
Cytoplasm |
PMID:9298646, PMID:11416161 |
| ||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MLMATIHVDG KEYEVNGADN LLEACLSLGL DIPYFCWHPA LGSVGACRQC AVKQYQNAED TRGRLVMSCM TPASDGTFIS IDDEEAKQFR ESVVEWLMTN HPHDCPVCEE GGNCHLQDMT VMTGHSFRRY RFTKRTHRNQ DLGPFISHEM NRCIACYRCV RYYKDYADGT DLGVYGAHDN VYFGRPEDGT LESEFSGNLV EICPTGVFTD KTHSERYNRK WDMQFAPSIC QQCSIGCNIS PGERYGELRR IENRYNGTVN HYFLCDRGRF GYGYVNLKDR PRQPVQRRGD DFITLNAEQA MQGAADILRQ SKKVIGIGSP RASVESNFAL RELVGEENFY TGIAHGEQER LQLALKVLRE GGIYTPALRE IESYDAVLVL GEDVTQTGAR VALAVRQAVK GKAREMAAAQ KVADWQIAAI LNIGQRAKHP LFVTNVDDTR LDDIAAWTYR APVEDQARLG FAIAHALDNS APAVDGIEPE LQSKIDVIVQ ALAGAKKPLI ISGTNAGSLE VIQAAANVAK ALKGRGADVG ITMIARSVNS MGLGIMGGGS LEEALTELET GRADAVVVLE NDLHRHASAI RVNAALAKAP LVMVVDHQRT AIMENAHLVL SAASFAESDG TVINNEGRAQ RFFQVYDPAY YDSKTVMLES WRWLHSLHST LLSREVDWTQ LDHVIDAVVA KIPELAGIKD AAPDATFRIR GQKLAREPHR YSGRTAMRAN ISVHEPRQPQ DIDTMFTFSM EGNNQPTAHR SQVPFAWAPG WNSPQAWNKF QDEVGGKLRF GDPGVRLFET SENGLDYFTS VPARFQPQDG KWRIAPYYHL FGSDELSQRA PVFQSRMPQP YIKLNPADAA KLGVNAGTRV SFSYDGNTVT LPVEIAEGLT AGQVGLPMGM SGIAPVLAGA HLEDLKEAQQ |
| Length |
910 |
| Mol. Wt |
100.543 kDa |
| pI |
6.2 (calculated) |
| Extinction coefficient |
109,210 - 111,210 (calc based on 29 Y, 12 W, and 16 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0007543 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG12087 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12087 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120001992 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2011 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
6.52E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
39.583+/-0.166 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.229685212 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
2657 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
1173 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
1547 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
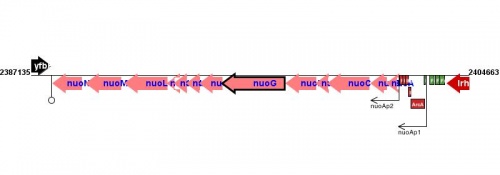 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:2398173..2398213
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for nuoG | |
|
microarray |
Summary of data for nuoG from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to nuoG Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG12087 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2011 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12087 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001992 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0007543 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Danio rerio |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Drosophila melanogaster |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Macaca mulatta |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Tetraodon nigroviridis |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
NUOG |
From SHIGELLACYC |
|
E. coli O157 |
NUOG |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
PF10588 NADH-ubiquinone oxidoreductase-G iron-sulfur binding region |
||
|
EcoCyc:EG12087 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12087 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001992 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2011 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0007543 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Bos taurus
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Canis familiaris
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Danio rerio
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Drosophila melanogaster
- Genes with homologs in Gallus gallus
- Genes with homologs in Homo sapiens
- Genes with homologs in Macaca mulatta
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Pan troglodytes
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Tetraodon nigroviridis
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


