lon:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
lon |
|---|---|
| Gene Synonym(s) |
ECK0433, b0439, JW0429, capR, deg, dir, muc, lopA[1], lonA, lopA |
| Product Desc. |
Component of DNA-binding, ATP-dependent protease La[3] DNA-binding, ATP-dependent protease LA; lon mutants form long cells[4] |
| Product Synonyms(s) |
DNA-binding ATP-dependent protease La[1], B0439[2][1], Muc[2][1], Dir[2][1], Deg[2][1], LopA[2][1], CapR[2][1], Lon[2][1] , capR, deg, dir, ECK0433, JW0429, lonA, lopA, muc, b0439 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Lon dependent lethality is due to induction of yefM-yoeb toxin/antitoxin pair.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
lon |
|---|---|
| Mnemonic |
Long form |
| Synonyms |
ECK0433, b0439, JW0429, capR, deg, dir, muc, lopA[1], lonA, lopA |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
9.87 minutes |
MG1655: 458112..460466 |
||
|
NC_012967: 430943..433297 |
||||
|
NC_012759: 360871..363225 |
||||
|
W3110 |
|
W3110: 458112..460466 |
||
|
DH10B: 397443..399797 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
458112 |
Edman degradation |
PMID:2984174 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
lon(del) (Keio:JW0429) |
deletion |
deletion |
PMID:16738554 |
||||
|
lon::Tn5KAN-2 (FB20175) |
Insertion at nt 1865 in Minus orientation |
PMID:15262929 |
does not contain pKD46 | ||||
|
lonK362A |
K362A |
Loss of proteolytic activity and ATP-binding |
seeded from UniProt:P0A9M0 | ||||
|
lonS679A |
S679A |
Loss of proteolytic activity |
seeded from UniProt:P0A9M0 | ||||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire Dextrin |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire D-Mannose |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire b-Methyl-D-glucoside |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire DL-a-Glycerol |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire D-Galactoniate |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire D-Galacturonate |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire D-Glucuronate |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire Acetate |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire a-Hydroxybutyrate |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire a-Ketobutyrate |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire a-Ketoglutarate |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire DL-Lactate |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire Succinate |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire Bromosuccinate |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire Glucuronamide |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire L-Alanine |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire L-Alanyl-glycine |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire L-Asparagine |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire L-Aspartate |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire Glycyl-L-aspartate |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire L-Serine |
PMID:16095938 |
|||
|
lon(del)::kan |
deletion |
Biolog:respiration |
unable to respire Uridine |
PMID:16095938 |
|||
|
lon-11 |
|||||||
|
lon-21 |
|||||||
|
lon-22 |
|||||||
|
lon-1::IS186 |
|||||||
|
lon-20 |
|||||||
|
lon-10 |
|||||||
|
lon-6 |
|||||||
|
lon-7 |
|||||||
|
lon-9 |
|||||||
|
lon-201 |
|||||||
|
lon-202 |
|||||||
|
lon-725(del)::kan |
PMID:16738554 |
||||||
|
capR9 |
|
PMID:17248613 |
|||||
|
capR9 |
derepression of UDPG pyrophosphorylase |
PMID:4910851 |
|||||
|
lon |
deletion |
Sensitivity to |
increases sensitivity to bicyclomycin |
PMID:21357484 |
fig 2 | ||
|
lon- |
Irradiated with UV light. |
Growth Phenotype |
Filament formation was reduced after being Irradiated. |
PMID 5335887 |
Strain AX14 was used. See Figure 1. | ||
|
Ion- |
Irradiation strain in YET agar plates. |
Colony Morphology |
Once exposed to UV radiation colony formation by the mutant cells was not observed. |
PMID:5335887 |
See Fig 3. D |
||
|
lon- |
Moved from YET broth to Glucose minimal agar |
Growth Phenotype |
The Lesions caused by the UV radiation were repaired. |
PMID:5335887 |
See Figure 4. | ||
|
lon- |
Puromycin addition |
Protein Synthesis |
Puromycin's inhibited L-arginine-UL-C incorporation by AX14 almost immediately, taking 25 minutes to complete |
PMID:5335887 |
See figure 6 | ||
|
lon725(del)::FRT |
deletion |
Mutagenesis |
Decrease in stress induced mutagenesis (SIM). |
PMID:23224554 |
Parent Strain: SMR4562 Experimental Strain: SMR12254 |
The mutation conferred a decrease in SIM with a reduction in mutant frequency by over 90 percent. See table S3 for full experimental data. | |
|
lon725(del)::FRT |
deletion |
Sensitivity to |
SDS-EDTA sensitivity |
PMID:23224554 |
Parent Strain: SMR4562 Experimental Strain: SMR12254 |
The mutation conferred increased sensitivity toward SDS-EDTA. See table S7 and S1 for summary of experimental data. | |
|
lon725(del)::FRT |
deletion |
Sensitivity to |
UV sensitivity |
PMID:23224554 |
Parent Strain: SMR4562 Experimental Strain: SMR12254 |
The mutation conferred increased sensitivity to UV. See table S7 and S1 for summary of experimental data. | |
|
CAG45114 lon725(del)::FRTKanFR |
deletion |
SigmaE activity |
decrease in sigmaE activity |
PMID:23224554 |
Parental Strain: CAG45114 Experimental Strain: SMR15273 |
See table S11 for full experimental results. | |
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW0429 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAATCCTGAGCGTTCTGAACG Primer 2:CCTTTTGCAGTCACAACCTGCAT | |
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 36% [6] | ||
|
Linked marker |
est. P1 cotransduction: 96% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10542 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10542 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000535 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0537 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0001521 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
Lon |
|---|---|
| Synonyms |
DNA-binding ATP-dependent protease La[1], B0439[2][1], Muc[2][1], Dir[2][1], Deg[2][1], LopA[2][1], CapR[2][1], Lon[2][1] , capR, deg, dir, ECK0433, JW0429, lonA, lopA, muc, b0439 |
| Product description |
Component of DNA-binding, ATP-dependent protease La[3] DNA-binding, ATP-dependent protease LA; lon mutants form long cells[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000166 |
nucleotide binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003593 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0238 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004176 |
ATP-dependent peptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001984 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004176 |
ATP-dependent peptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003111 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004176 |
ATP-dependent peptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003111 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004176 |
ATP-dependent peptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004815 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004176 |
ATP-dependent peptidase activity |
PMID:6458036 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0004176 |
ATP-dependent peptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008268 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
PMID:15037242 |
IPI: Inferred from Physical Interaction |
F |
Missing: with/from | |||
|
GO:0004176 |
ATP-dependent peptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008269 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016887 |
ATPase activity |
PMID:6458036 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0004252 |
serine-type endopeptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001984 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008233 |
peptidase activity |
PMID:9720920 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0004252 |
serine-type endopeptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008268 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006508 |
proteolysis |
PMID:6458036 |
IDA: Inferred from Direct Assay |
P |
complete | |||
|
GO:0004176 |
ATP-dependent peptidase activity |
PMID:9720920 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0004252 |
serine-type endopeptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008269 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
PMID:8995294 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003959 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
PMID:8995294 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004815 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006950 |
response to stress |
PMID:6330035 |
IEP: Inferred from Expression Pattern |
P |
complete | |||
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004252 |
serine-type endopeptidase activity |
PMID:15606774 |
IMP: Inferred from Mutant Phenotype |
F |
complete | |||
|
GO:0005737 |
cytoplasm |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0963 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009408 |
response to heat |
PMID:8349564 |
IEP: Inferred from Expression Pattern |
P |
complete | |||
|
GO:0005737 |
cytoplasm |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0086 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006508 |
proteolysis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001984 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006508 |
proteolysis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003111 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006508 |
proteolysis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004815 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006508 |
proteolysis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008268 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006508 |
proteolysis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008269 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006950 |
response to stress |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0346 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of DNA-binding, ATP-dependent protease La |
could be indirect |
||
|
Protein |
ssb |
PMID:15690043 |
Experiment(s):EBI-894571 | |
|
Protein |
rplC |
PMID:15690043 |
Experiment(s):EBI-889577 | |
|
Protein |
rplI |
PMID:15690043 |
Experiment(s):EBI-889577, EBI-894571 | |
|
Protein |
rplM |
PMID:15690043 |
Experiment(s):EBI-889577, EBI-894571 | |
|
Protein |
rplT |
PMID:15690043 |
Experiment(s):EBI-889577 | |
|
Protein |
rpsB |
PMID:15690043 |
Experiment(s):EBI-889577 | |
|
Protein |
rpsC |
PMID:15690043 |
Experiment(s):EBI-889577 | |
|
Protein |
tufA |
PMID:15690043 |
Experiment(s):EBI-889577 | |
|
Protein |
fsaA |
PMID:15690043 |
Experiment(s):EBI-894571 | |
|
Protein |
rplU |
PMID:15690043 |
Experiment(s):EBI-894571 | |
|
Protein |
rplV |
PMID:15690043 |
Experiment(s):EBI-894571 | |
|
Protein |
rpsG |
PMID:15690043 |
Experiment(s):EBI-894571 | |
|
Protein |
rpsH |
PMID:15690043 |
Experiment(s):EBI-894571 | |
|
Protein |
yjgL |
PMID:15690043 |
Experiment(s):EBI-894571 | |
|
Protein |
dnaX |
PMID:16606699 |
Experiment(s):EBI-1136659 | |
|
Protein |
sapF |
PMID:16606699 |
Experiment(s):EBI-1136659 | |
|
Protein |
secB |
PMID:16606699 |
Experiment(s):EBI-1136659 | |
|
Protein |
dnaA |
PMID:16606699 |
Experiment(s):EBI-1136659 | |
|
Protein |
nadE |
PMID:16606699 |
Experiment(s):EBI-1136659 | |
|
Protein |
ssb |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):2.272167 | |
|
Protein |
rpsC |
PMID:19402753 |
MALDI(Z-score):31.921524 | |
|
Protein |
rplI |
PMID:19402753 |
LCMS(ID Probability):99.0 MALDI(Z-score):17.053160 | |
|
Protein |
rplU |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
yjgL |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplM |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):28.085510 | |
|
Protein |
rplA |
PMID:19402753 |
MALDI(Z-score):23.969367 | |
|
Protein |
clpA |
PMID:19402753 |
MALDI(Z-score):22.779291 | |
|
Protein |
tufB |
PMID:19402753 |
MALDI(Z-score):22.916243 | |
|
Small Molecule |
Diisopropyl fluorophosphate |
PMID:6458037 |
||
|
Small Molecule |
Mg2+ |
PMID:6458037 |
Data was not shown, but indicated that the presence of both Mg2+ and ATP were required for proteolytic activity. | |
|
Small Molecule |
ATP |
PMID:6458037 |
See evidence for Mg2+ | |
| edit table |
</protect>
Notes
For a review of Lon and other proteases, see Proteases and their targets in Escherichia coli. Gottesman (1996)[7]
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MNPERSERIE IPVLPLRDVV VYPHMVIPLF VGREKSIRCL EAAMDHDKKI MLVAQKEAST DEPGVNDLFT VGTVASILQM LKLPDGTVKV LVEGLQRARI SALSDNGEHF SAKAEYLESP TIDEREQEVL VRTAISQFEG YIKLNKKIPP EVLTSLNSID DPARLADTIA AHMPLKLADK QSVLEMSDVN ERLEYLMAMM ESEIDLLQVE KRIRNRVKKQ MEKSQREYYL NEQMKAIQKE LGEMDDAPDE NEALKRKIDA AKMPKEAKEK AEAELQKLKM MSPMSAEATV VRGYIDWMVQ VPWNARSKVK KDLRQAQEIL DTDHYGLERV KDRILEYLAV QSRVNKIKGP ILCLVGPPGV GKTSLGQSIA KATGRKYVRM ALGGVRDEAE IRGHRRTYIG SMPGKLIQKM AKVGVKNPLF LLDEIDKMSS DMRGDPASAL LEVLDPEQNV AFSDHYLEVD YDLSDVMFVA TSNSMNIPAP LLDRMEVIRL SGYTEDEKLN IAKRHLLPKQ IERNALKKGE LTVDDSAIIG IIRYYTREAG VRGLEREISK LCRKAVKQLL LDKSLKHIEI NGDNLHDYLG VQRFDYGRAD NENRVGQVTG LAWTEVGGDL LTIETACVPG KGKLTYTGSL GEVMQESIQA ALTVVRARAE KLGINPDFYE KRDIHVHVPE GATPKDGPSA GIAMCTALVS CLTGNPVRAD VAMTGEITLR GQVLPIGGLK EKLLAAHRGG IKTVLIPFEN KRDLEEIPDN VIADLDIHPV KRIEEVLTLA LQNEPSGMQV VTAK |
| Length |
784 |
| Mol. Wt |
87.44 kDa |
| pI |
6.2 (calculated) |
| Extinction coefficient |
46,300 - 47,050 (calc based on 20 Y, 3 W, and 6 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0001521 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10542 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10542 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000535 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0537 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
1.55E+03 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
158.851+/-0.689 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.11108+/-0.01107 |
Molecules/cell |
|
by FISH |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.204757859 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
5477 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
1485 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
4688 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
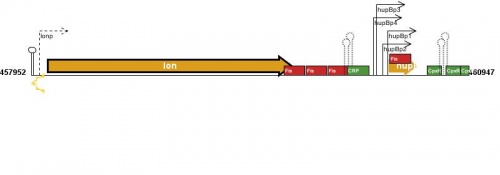 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:458092..458132
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for lon | |
|
microarray |
Summary of data for lon from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (457843..458176) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to lon Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10542 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0537 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10542 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000535 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0001521 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Danio rerio |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Drosophila melanogaster |
|
From Inparanoid:20070104 |
|
Drosophila pseudoobscura |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Macaca mulatta |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Saccharomyces cerevisiae |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Tetraodon nigroviridis |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
LON |
From SHIGELLACYC |
|
E. coli O157 |
LON |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
PF00004 ATPase family associated with various cellular activities (AAA) |
||
|
EcoCyc:EG10542 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10542 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000535 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0537 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0001521 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Gottesman, S (1996) Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30 465-506 PubMed
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories
- Genes in OpenBioSystems with Promoter Fusions
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Apis mellifera
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Bos taurus
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Canis familiaris
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Danio rerio
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Drosophila melanogaster
- Genes with homologs in Drosophila pseudoobscura
- Genes with homologs in Gallus gallus
- Genes with homologs in Homo sapiens
- Genes with homologs in Macaca mulatta
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Oryza gramene
- Genes with homologs in Pan troglodytes
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Saccharomyces cerevisiae
- Genes with homologs in Schizosaccharomyces pombe
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Tetraodon nigroviridis
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


