sbcC:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
sbcC |
|---|---|
| Gene Synonym(s) |
ECK0391, b0397, JW0387, rmuA[1], rmuA |
| Product Desc. |
ATP-dependent dsDNA exonuclease[2][3]; Component of SbcCD ATP-dependent dsDNA exonuclease[3] DNA hairpin dsDNA 3'-exonuclease SbcCD, Mn(2+), ATP-dependent; ATP-independent 5' ssDNA endonuclease; required for recombinational repair of dsDNA breaks; cosuppressor with sbcB of recB recC mutations; heterodimeric[4] |
| Product Synonyms(s) |
exonuclease, dsDNA, ATP-dependent[1], B0397[2][1], SbcC[2][1] , ECK0391, JW0387, rmuA, b0397 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
SbcCD can cleave dsDNA 5' ends sealed by a hairpin or by covalent linko a protein. SbcCD limits recombination between short inverted repeats, but is also reported to be required for palindrome-stimulated deletion. SbcCD protein forms a large 1.2 megadalton complex. Second amino acid was sequenced in protein as Gly, but in all DNA samples is a Lys codon. Coiled coil protein.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
sbcC |
|---|---|
| Mnemonic |
Suppression of recBC |
| Synonyms |
ECK0391, b0397, JW0387, rmuA[1], rmuA |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
8.88 minutes |
MG1655: 414977..411831 |
||
|
NC_012967: 384176..381030 |
||||
|
NC_012759: 314590..317736 |
||||
|
W3110 |
|
W3110: 414977..411831 |
||
|
DH10B: 354308..351162 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
411831 |
Edman degradation |
PMID:9242643 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
sbcC(del) (Keio:JW0387) |
deletion |
deletion |
PMID:16738554 |
||||
|
sbcC761(del)::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW0387 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAAAATTCTCAGCCTGCGCCT Primer 2:CCTTTCACTGCAAACGTACTTTC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 54% [6] | ||
|
Linked marker |
est. P1 cotransduction: 49% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10927 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10927 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000916 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0920 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0001381 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
SbcC |
|---|---|
| Synonyms |
exonuclease, dsDNA, ATP-dependent[1], B0397[2][1], SbcC[2][1] , ECK0391, JW0387, rmuA, b0397 |
| Product description |
ATP-dependent dsDNA exonuclease[2][3]; Component of SbcCD ATP-dependent dsDNA exonuclease[3] DNA hairpin dsDNA 3'-exonuclease SbcCD, Mn(2+), ATP-dependent; ATP-independent 5' ssDNA endonuclease; required for recombinational repair of dsDNA breaks; cosuppressor with sbcB of recB recC mutations; heterodimeric[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004518 |
nuclease activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0540 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004519 |
endonuclease activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0255 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004527 |
exonuclease activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004592 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004527 |
exonuclease activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0269 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006259 |
DNA metabolic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004592 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006260 |
DNA replication |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0235 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006310 |
DNA recombination |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0233 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016787 |
hydrolase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0378 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of SbcCD ATP-dependent dsDNA exonuclease |
could be indirect |
||
|
Protein |
ynbC |
PMID:15690043 |
Experiment(s):EBI-879066 | |
|
Protein |
prs |
PMID:16606699 |
Experiment(s):EBI-1136452 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MKILSLRLKN LNSLKGEWKI DFTREPFASN GLFAITGPTG AGKTTLLDAI CLALYHETPR LSNVSQSQND LMTRDTAECL AEVEFEVKGE AYRAFWSQNR ARNQPDGNLQ VPRVELARCA DGKILADKVK DKLELTATLT GLDYGRFTRS MLLSQGQFAA FLNAKPKERA ELLEELTGTE IYGQISAMVF EQHKSARTEL EKLQAQASGV TLLTPEQVQS LTASLQVLTD EEKQLITAQQ QEQQSLNWLT RQDELQQEAS RRQQALQQAL AEEEKAQPQL AALSLAQPAR NLRPHWERIA EHSAALAHIR QQIEEVNTRL QSTMALRASI RHHAAKQSAE LQQQQQSLNT WLQEHDRFRQ WNNEPAGWRA QFSQQTSDRE HLRQWQQQLT HAEQKLNALA AITLTLTADE VATALAQHAE QRPLRQHLVA LHGQIVPQQK RLAQLQVAIQ NVTQEQTQRN AALNEMRQRY KEKTQQLADV KTICEQEARI KTLEAQRAQL QAGQPCPLCG STSHPAVEAY QALEPGVNQS RLLALENEVK KLGEEGATLR GQLDAITKQL QRDENEAQSL RQDEQALTQQ WQAVTASLNI TLQPLDDIQP WLDAQDEHER QLRLLSQRHE LQGQIAAHNQ QIIQYQQQIE QRQQLLLTTL TGYALTLPQE DEEESWLATR QQEAQSWQQR QNELTALQNR IQQLTPILET LPQSDELPHC EETVVLENWR QVHEQCLALH SQQQTLQQQD VLAAQSLQKA QAQFDTALQA SVFDDQQAFL AALMDEQTLT QLEQLKQNLE NQRRQAQTLV TQTAETLAQH QQHRPDDGLA LTVTVEQIQQ ELAQTHQKLR ENTTSQGEIR QQLKQDADNR QQQQTLMQQI AQMTQQVEDW GYLNSLIGSK EGDKFRKFAQ GLTLDNLVHL ANQQLTRLHG RYLLQRKASE ALEVEVVDTW QADAVRDTRT LSGGESFLVS LALALALSDL VSHKTRIDSL FLDEGFGTLD SETLDTALDA LDALNASGKT IGVISHVEAM KERIPVQIKV KKINGLGYSK LESTFAVK |
| Length |
1,048 |
| Mol. Wt |
118.72 kDa |
| pI |
5.5 (calculated) |
| Extinction coefficient |
98,890 - 99,890 (calc based on 11 Y, 15 W, and 8 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0001381 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10927 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10927 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000916 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0920 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
Ecoli K-12 |
3.02+/-0.066 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.008423395 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
135 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
59 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
66 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
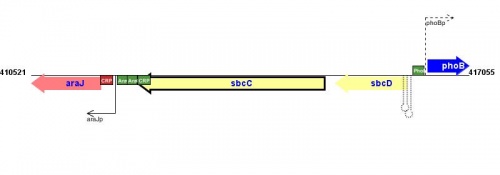 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:414957..414997
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for sbcC | |
|
microarray |
Summary of data for sbcC from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to sbcC Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10927 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0920 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10927 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000916 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0001381 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Danio rerio |
|
From Inparanoid:20070104 |
|
Drosophila melanogaster |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Saccharomyces cerevisiae |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
SBCC |
From SHIGELLACYC |
|
E. coli O157 |
SBCC |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10927 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10927 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000916 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0920 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0001381 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Danio rerio
- Genes with homologs in Drosophila melanogaster
- Genes with homologs in Gallus gallus
- Genes with homologs in Mus musculus
- Genes with homologs in Oryza gramene
- Genes with homologs in Pan troglodytes
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Saccharomyces cerevisiae
- Genes with homologs in Schizosaccharomyces pombe
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


