rpoD:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
rpoD |
|---|---|
| Gene Synonym(s) |
ECK3057, b3067, JW3039, alt[1], alt |
| Product Desc. |
sigma70 factor[2]; RNA polymerase, sigma 70 (sigma D) factor[3]; Component of RNA polymerase sigma70[2]; RNA polymerase sigma 70[3] RNA polymerase subunit, sigma 70, initiates transcription; housekeeping sigma[4] |
| Product Synonyms(s) |
RNA polymerase, sigma 70 (sigma D) factor[1], B3067[2][1], Alt[2][1], RpoD[2][1], sigmaD factor[2][1], sigma D factor[3][1], sigma 70 factor[3][1] , alt, ECK3057, JW3039, b3067 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression |
transcription unit(s): rpoD[2], rpsU-dnaG-rpoD[2] |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
rpoD |
|---|---|
| Mnemonic |
RNA polymerase |
| Synonyms |
ECK3057, b3067, JW3039, alt[1], alt |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
69.21 minutes |
MG1655: 3211069..3212910 |
||
|
NC_012967: 3145864..3147705 |
||||
|
NC_012759: 3098217..3100058 |
||||
|
W3110 |
|
W3110: 3211703..3213544 |
||
|
DH10B: 3308814..3310655 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
3211069 |
Edman degradation |
PMID:1095419 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
rpoD800 |
Growth Phenotype |
Temperature-sensitive for growth at 42°C. |
|||||
|
rpoD2 (alt) |
R596H |
PMID:3889551 PMID:351406 |
|||||
|
rpoD40 |
Growth Phenotype |
Nonsense mutation |
PMID:7007812 |
||||
|
rpoD(P504L) |
P504L |
Suppresses multiple amino acid auxotrophy of relA spoT double mutant |
PMID:7563072 |
||||
|
rpoD(S506F) |
S506F |
Suppresses multiple amino acid auxotrophy of relA spoT double mutant |
PMID:7563072 |
||||
|
rpoD800(ts) |
deletion 329-342 |
temperature sensitive |
|||||
|
rpoD285(ts) |
deletion 329-342 |
temperature sensitive |
PMID:2841288 PMID:3052291 PMID:366614 PMID:6355770 |
||||
|
rpoD899(ts) |
temperature sensitive |
||||||
|
rpoD(K99E) |
K99E |
defective in abortive initiation | |||||
| edit table |
<protect></protect>
Notes
The Keio collection[5] lists a deletion of rpoD, which is an essential gene. The insertion in this strain is a duplication of the rpoD region.[6]
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW3039 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCGAGCAAAACCCGCAGTCACA Primer 2:CCATCGTCCAGGAAGCTACGCAG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 27% [8] | ||
|
Linked marker |
est. P1 cotransduction: 87% [8] Synonyms:zgh-3075::Tn10, zgj-3075::Tn10 | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10896 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10896 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000887 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0889 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010070 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
RpoD |
|---|---|
| Synonyms |
RNA polymerase, sigma 70 (sigma D) factor[1], B3067[2][1], Alt[2][1], RpoD[2][1], sigmaD factor[2][1], sigma D factor[3][1], sigma 70 factor[3][1] , alt, ECK3057, JW3039, b3067 |
| Product description |
sigma70 factor[2]; RNA polymerase, sigma 70 (sigma D) factor[3]; Component of RNA polymerase sigma70[2]; RNA polymerase sigma 70[3] RNA polymerase subunit, sigma 70, initiates transcription; housekeeping sigma[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0016987 |
sigma factor activity |
PMID:4882047 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000943 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007127 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007624 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007627 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007630 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007631 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR009042 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012760 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009408 |
response to heat |
PMID:8349564 |
IEP: Inferred from Expression Pattern |
P |
complete | |||
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013324 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013325 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR014284 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0238 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003700 |
transcription factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000943 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003700 |
transcription factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007624 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003700 |
transcription factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007627 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003700 |
transcription factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007630 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003700 |
transcription factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007631 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003700 |
transcription factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR009042 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003700 |
transcription factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013324 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003700 |
transcription factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013325 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003700 |
transcription factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR014284 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005515 |
protein binding |
PMID:10764785 |
IPI: Inferred from Physical Interaction |
UniProtKB:P0A8T7 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006350 |
transcription |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0804 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006352 |
transcription initiation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000943 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006352 |
transcription initiation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007624 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006352 |
transcription initiation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007627 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006352 |
transcription initiation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007630 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006352 |
transcription initiation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007631 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006352 |
transcription initiation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR009042 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006352 |
transcription initiation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012760 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006352 |
transcription initiation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013324 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006352 |
transcription initiation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013325 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006352 |
transcription initiation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR014284 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006355 |
regulation of transcription, DNA-dependent |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000943 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006355 |
regulation of transcription, DNA-dependent |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007624 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006355 |
regulation of transcription, DNA-dependent |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007627 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006355 |
regulation of transcription, DNA-dependent |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007630 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006355 |
regulation of transcription, DNA-dependent |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007631 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006355 |
regulation of transcription, DNA-dependent |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR009042 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006355 |
regulation of transcription, DNA-dependent |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012760 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006355 |
regulation of transcription, DNA-dependent |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013324 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006355 |
regulation of transcription, DNA-dependent |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013325 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006355 |
regulation of transcription, DNA-dependent |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR014284 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006355 |
regulation of transcription, DNA-dependent |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0731 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016987 |
sigma factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000943 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016987 |
sigma factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007624 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016987 |
sigma factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007627 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016987 |
sigma factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007630 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016987 |
sigma factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007631 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016987 |
sigma factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR009042 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016987 |
sigma factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012760 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016987 |
sigma factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013324 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016987 |
sigma factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013325 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016987 |
sigma factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR014284 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016987 |
sigma factor activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0731 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030528 |
transcription regulator activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007127 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0045449 |
regulation of transcription |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007127 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0045449 |
regulation of transcription |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0805 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of RNA polymerase sigma70 |
could be indirect |
||
|
Protein |
rpoC |
PMID:15690043 |
Experiment(s):EBI-886257, EBI-879261, EBI-883046, EBI-892741 | |
|
Protein |
rpoZ |
PMID:15690043 |
Experiment(s):EBI-886257, EBI-879161, EBI-882858, EBI-892741 | |
|
Protein |
rpsB |
PMID:15690043 |
Experiment(s):EBI-886257 | |
|
Protein |
rpsC |
PMID:15690043 |
Experiment(s):EBI-886257 | |
|
Protein |
rpsD |
PMID:15690043 |
Experiment(s):EBI-886257 | |
|
Protein |
rpsE |
PMID:15690043 |
Experiment(s):EBI-886257 | |
|
Protein |
rpsG |
PMID:15690043 |
Experiment(s):EBI-886257, EBI-892741 | |
|
Protein |
accA |
PMID:15690043 |
Experiment(s):EBI-886257 | |
|
Protein |
infB |
PMID:15690043 |
Experiment(s):EBI-886257, EBI-892741 | |
|
Protein |
nusA |
PMID:15690043 |
Experiment(s):EBI-886257, EBI-892741, EBI-893117 | |
|
Protein |
rplA |
PMID:15690043 |
Experiment(s):EBI-886257, EBI-892741 | |
|
Protein |
rplB |
PMID:15690043 |
Experiment(s):EBI-886257 | |
|
Protein |
rplD |
PMID:15690043 |
Experiment(s):EBI-886257, EBI-892741 | |
|
Protein |
rplO |
PMID:15690043 |
Experiment(s):EBI-886257 | |
|
Protein |
rplV |
PMID:15690043 |
Experiment(s):EBI-886257 | |
|
Protein |
rpoA |
PMID:15690043 |
Experiment(s):EBI-886257, EBI-879354, EBI-883271, EBI-892741 | |
|
Protein |
rpoB |
PMID:15690043 |
Experiment(s):EBI-886257, EBI-879304, EBI-883170, EBI-892741 | |
|
Protein |
pepB |
PMID:15690043 |
Experiment(s):EBI-892741 | |
|
Protein |
rho |
PMID:15690043 |
Experiment(s):EBI-892741 | |
|
Protein |
rplL |
PMID:15690043 |
Experiment(s):EBI-892741 | |
|
Protein |
rplS |
PMID:15690043 |
Experiment(s):EBI-892741 | |
|
Protein |
rplU |
PMID:15690043 |
Experiment(s):EBI-892741 | |
|
Protein |
rpmB |
PMID:15690043 |
Experiment(s):EBI-892741 | |
|
Protein |
rpsA |
PMID:15690043 |
Experiment(s):EBI-892741 | |
|
Protein |
rpsF |
PMID:15690043 |
Experiment(s):EBI-892741 | |
|
Protein |
tufA |
PMID:15690043 |
Experiment(s):EBI-892741 | |
|
Protein |
yeeX |
PMID:15690043 |
Experiment(s):EBI-892741 | |
|
Protein |
rho |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
rpoB |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
dnaG |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
nadE |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
rplE |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
matC |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
potD |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
rpsB |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
rpoC |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
mfd |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
yfaS |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
yniC |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
gadX |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
rplD |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
gmhB |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
blc |
PMID:16606699 |
Experiment(s):EBI-1144702 | |
|
Protein |
pepB |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rpmB |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplU |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
infB |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):10.756318 | |
|
Protein |
rpsF |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rpsA |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):6.241428 | |
|
Protein |
rho |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplS |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplA |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):29.174139 | |
|
Protein |
rplD |
PMID:19402753 |
LCMS(ID Probability):99.4 MALDI(Z-score):17.053160 | |
|
Protein |
rpoZ |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):27.779532 | |
|
Protein |
nusA |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):23.202327 | |
|
Protein |
rpoB |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):26.899873 | |
|
Protein |
rpoC |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):14.555460 | |
|
Protein |
yeeX |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplL |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MEQNPQSQLK LLVTRGKEQG YLTYAEVNDH LPEDIVDSDQ IEDIIQMIND MGIQVMEEAP DADDLMLAEN TADEDAAEAA AQVLSSVESE IGRTTDPVRM YMREMGTVEL LTREGEIDIA KRIEDGINQV QCSVAEYPEA ITYLLEQYDR VEAEEARLSD LITGFVDPNA EEDLAPTATH VGSELSQEDL DDDEDEDEED GDDDSADDDN SIDPELAREK FAELRAQYVV TRDTIKAKGR SHATAQEEIL KLSEVFKQFR LVPKQFDYLV NSMRVMMDRV RTQERLIMKL CVEQCKMPKK NFITLFTGNE TSDTWFNAAI AMNKPWSEKL HDVSEEVHRA LQKLQQIEEE TGLTIEQVKD INRRMSIGEA KARRAKKEMV EANLRLVISI AKKYTNRGLQ FLDLIQEGNI GLMKAVDKFE YRRGYKFSTY ATWWIRQAIT RSIADQARTI RIPVHMIETI NKLNRISRQM LQEMGREPTP EELAERMLMP EDKIRKVLKI AKEPISMETP IGDDEDSHLG DFIEDTTLEL PLDSATTESL RAATHDVLAG LTAREAKVLR MRFGIDMNTD YTLEEVGKQF DVTRERIRQI EAKALRKLRH PSRSEVLRSF LDD |
| Length |
613 |
| Mol. Wt |
70.264 kDa |
| pI |
4.5 (calculated) |
| Extinction coefficient |
41,370 - 41,745 (calc based on 13 Y, 4 W, and 3 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0010070 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10896 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10896 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000887 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0889 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
1.32E+03 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
E. coli K-12 MG1655 |
11809 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
2284 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
5289 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
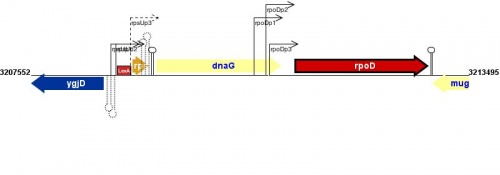 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:3211049..3211089
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for rpoD | |
|
microarray |
Summary of data for rpoD from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (3210420..3210769) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to rpoD Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10896 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0889 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10896 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000887 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010070 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
RPOD |
From SHIGELLACYC |
|
E. coli O157 |
RPOD |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10896 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10896 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000887 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0889 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010070 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ Baba, T et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2 2006.0008 PubMed
- ↑ Yamamoto, N et al. (2009) Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 5 335 PubMed
- ↑ 7.0 7.1 CGSC: The Coli Genetics Stock Center
- ↑ 8.0 8.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories


