rho:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
rho |
|---|---|
| Gene Synonym(s) |
ECK3775, b3783, JW3756, nitA, nusD, psuA, rnsC, sun, tsu, psu, sbaA, suA[1], hdf, tabC |
| Product Desc. |
transcription termination factor Rho; polarity suppressor[2][3]; Component of transcription termination factor Rho[3] Transcription termination factor Rho; bicyclomycin target[4] |
| Product Synonyms(s) |
transcription termination factor[1], B3783[2][1], RnsC[2][1], Tsu[2][1], Sun[2][1], NusD[2][1], Psu[2][1], SbaA[2][1], PsuA[2][1], NitA[2][1], Rho[2][1] , ECK3775, hdf, JW3756, nitA, nusD, psuA, rnsC, sbaA, sun, tabC, tsu, b3783 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
- The rho gene is often misannotated as a polarity suppressor. Mutations in rho suppress polarity.[5][6][7][8][9] Wild type rho is the source of polarity.
- rho has been shown to be an essential gene (Das, 1976; Bubunenko, 2007).[4]
- rho is required to maintain genomic integrity by resolving transcription/replication conflicts [10]
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ Das, A et al. (1976) Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc. Natl. Acad. Sci. U.S.A. 73 1959-63 PubMed
- ↑ Korn, LJ & Yanofsky, C (1976) Polarity suppressors defective in transcription termination at the attenuator of the tryptophan operon of Escherichia coli have altered rho factor. J. Mol. Biol. 106 231-41 PubMed
- ↑ Belfort, M & Oppenheim, AB (1976) Efficient suppression of the requirement for N function of bacteriophage lambda by a Rho-defective E.coli suA mutant. Mol. Gen. Genet. 148 171-3 PubMed
- ↑ Richardson, JP & Carey, JL 3rd (1982) rho Factors from polarity suppressor mutants with defects in their RNA interactions. J. Biol. Chem. 257 5767-71 PubMed
- ↑ Das, A et al. (1978) Interaction of RNA polymerase and rho in transcription termination: coupled ATPase. Proc. Natl. Acad. Sci. U.S.A. 75 4828-32 PubMed
- ↑ Washburn, RS & Gottesman, ME (2011) Transcription termination maintains chromosome integrity. Proc. Natl. Acad. Sci. U.S.A. 108 792-7 PubMed
</noinclude>
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
rho |
|---|---|
| Mnemonic |
Rho termination factor |
| Synonyms |
ECK3775, b3783, JW3756, nitA, nusD, psuA, rnsC, sun, tsu, psu, sbaA, suA[1], hdf, tabC |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
85.45 minutes |
MG1655: 3964440..3965699 |
||
|
NC_012967: 3928217..3929476 |
||||
|
NC_012759: 3854109..3855368 |
||||
|
W3110 |
|
W3110: 3670264..3669005 |
||
|
DH10B: 4063360..4064619 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
3964440 |
Edman degradation |
PMID:6219230 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
rhoF64L,A |
F64L,A |
Defective for RNA-binding |
seeded from UniProt:P0AG30 | ||||
|
rhoK181Q |
K181Q |
Partial loss of ATPase, helicase and termination activity |
seeded from UniProt:P0AG30 | ||||
|
rhoK184Q |
K184Q |
Improves ATPase and helicase activity but reduced termination activity |
seeded from UniProt:P0AG30 | ||||
|
rhoF62L,A |
F62L,A |
Defective for RNA-binding |
seeded from UniProt:P0AG30 | ||||
|
rhoD265N |
D265N |
Loss of ATPase activity, helicase and termination activity |
seeded from UniProt:P0AG30 | ||||
|
rho-15(Ts) |
IS1 insertion into C-terminus |
changes seven amino acids, eliminates the last two (changes DFFEMMKRS to EVMTPTY) |
Growth Phenotype |
fails to grow at 42oC, grows slowly at 30oC, suppresses polarity |
quickly loses temperature-sensitive phenotype due to accumulation of suppressor mutations.[4] | ||
|
rho-4 |
C to A transversion in codon 243 |
A243E |
polarity suppression |
suppresses polarity, defective for termination at Rho-dependent terminators |
Originally thought to be an amber (UAG) mutation,[5] but later found to be a missense mutation.[6][7] | ||
|
rho-112(Am) |
amber (UAG) mutation | ||||||
|
rho-702(ts) |
Growth Phenotype |
temperature sensitive |
|||||
|
rho-6 |
|||||||
|
rho026 (aka rhoHDF026 |
Increased degradation of abnormal proteins, such as puromycyl polypeptides or canavanine-containing polypeptides. |
PMID:377279 |
|||||
|
rho026 (aka rhoHDF026) |
P103L, S153Y |
Unable to support growth of phage T4 and inhibits Lambda phage N activity. Uninfected cells were temperature-sensitive for growth at 42°C in original isolate.
|
PMID:6999171 PMID:6225121 PMID:8757797 |
P103L change is responsible for interfering with T4 plating and Lambda N-mediated antitermination. S153Y was acquired as a suppressor of ts phenotype. | |||
|
sbaA mutation in SDT1 |
deletion |
Growth Phenotype |
deletion of sbaA gene causes limited growth at higher temperatures. |
PMID:6247616 |
See fig 2B for gel electrophoresis results. | ||
|
sbaA mutation in SDT1 |
deletion |
metabolism |
Increase in the accumulation of pppGpp |
PMID:6247616 |
See Fig 3 A & B for full experimental results. | ||
|
sbaA mutation in SDT1 |
deletion |
beta-galactosidase activity |
decrease in the beta-galactosidase activity |
PMID:6247616 |
See Fig 4 for full results. | ||
|
sbaA mutation in SDT1 |
deletion |
beta-galactosidase activity |
addition of 1.0 mM cAMP causes increased beta-galactosidase activity but was significantly lower than parent strain. |
PMID:6247616 |
See Fig 4 for full results. | ||
| edit table |
<protect></protect>
Notes
The Keio deletion appears to be the result of a gene duplication, wt rho is still present in the strain.[8]
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW3756 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAATCTTACCGAATTAAAGAA Primer 2:CCTGAGCGTTTCATCATTTCGAA | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 67% [10] | ||
|
metEo-3079::Tn10 |
Linked marker |
est. P1 cotransduction: 14% [10] | |
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10845 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10845 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000836 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0838 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012362 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
Rho |
|---|---|
| Synonyms |
transcription termination factor[1], B3783[11][1], RnsC[11][1], Tsu[11][1], Sun[11][1], NusD[11][1], Psu[11][1], SbaA[11][1], PsuA[11][1], NitA[11][1], Rho[11][1] , ECK3775, hdf, JW3756, nitA, nusD, psuA, rnsC, sbaA, sun, tabC, tsu, b3783 |
| Product description |
transcription termination factor Rho; polarity suppressor[11][12]; Component of transcription termination factor Rho[12] Transcription termination factor Rho; bicyclomycin target[13] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000166 |
nucleotide binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003593 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003676 |
nucleic acid binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011129 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003715 |
transcription termination factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004665 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003715 |
transcription termination factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011112 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003715 |
transcription termination factor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011113 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003723 |
RNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011113 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003723 |
RNA binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0694 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004386 |
helicase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0347 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004665 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006350 |
transcription |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0804 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006353 |
transcription termination |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004665 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006353 |
transcription termination |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011112 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006353 |
transcription termination |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011113 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006353 |
transcription termination |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0806 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016787 |
hydrolase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0378 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0017111 |
nucleoside-triphosphatase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003593 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0031555 |
transcriptional attenuation |
PMID:2423505 |
IMP: Inferred from Mutant Phenotype |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of transcription termination factor Rho |
could be indirect |
||
|
Protein |
tufA |
PMID:15690043 |
Experiment(s):EBI-884347 | |
|
Protein |
gsp |
PMID:15690043 |
Experiment(s):EBI-891277 | |
|
Protein |
menF |
PMID:15690043 |
Experiment(s):EBI-891277 | |
|
Protein |
rplC |
PMID:15690043 |
Experiment(s):EBI-891277 | |
|
Protein |
rplL |
PMID:15690043 |
Experiment(s):EBI-891277 | |
|
Protein |
rpsB |
PMID:15690043 |
Experiment(s):EBI-891277 | |
|
Protein |
yhbY |
PMID:15690043 |
Experiment(s):EBI-891277 | |
|
Protein |
uspG |
PMID:16606699 |
Experiment(s):EBI-1146678 | |
|
Protein |
yhbY |
PMID:19402753 |
LCMS(ID Probability):99.0 MALDI(Z-score):27.463697 | |
|
Protein |
tufB |
PMID:19402753 |
MALDI(Z-score):20.423130 | |
|
Protein |
rplD |
PMID:19402753 |
LCMS(ID Probability):99.0 MALDI(Z-score):17.053160 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MNLTELKNTP VSELITLGEN MGLENLARMR KQDIIFAILK QHAKSGEDIF GDGVLEILQD GFGFLRSADS SYLAGPDDIY VSPSQIRRFN LRTGDTISGK IRPPKEGERY FALLKVNEVN FDKPENARNK ILFENLTPLH ANSRLRMERG NGSTEDLTAR VLDLASPIGR GQRGLIVAPP KAGKTMLLQN IAQSIAYNHP DCVLMVLLID ERPEEVTEMQ RLVKGEVVAS TFDEPASRHV QVAEMVIEKA KRLVEHKKDV IILLDSITRL ARAYNTVVPA SGKVLTGGVD ANALHRPKRF FGAARNVEEG GSLTIIATAL IDTGSKMDEV IYEEFKGTGN MELHLSRKIA EKRVFPAIDY NRSGTRKEEL LTTQEELQKM WILRKIIHPM GEIDAMEFLI NKLAMTKTND DFFEMMKRS |
| Length |
419 |
| Mol. Wt |
47.005 kDa |
| pI |
7.2 (calculated) |
| Extinction coefficient |
15,930 - 16,055 (calc based on 7 Y, 1 W, and 1 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
|
Chemical inhibitor of Rho | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0012362 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10845 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10845 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000836 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0838 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
4.45E+03 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
2165.11+/-23.867 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.250595711 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 EMG2 |
300 |
molecules/cell |
|
Quantitative protein sequencing |
Spot ID: M68-l |
PMID: 9298646 |
|
Protein |
E. coli K-12 MG1655 |
17921 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
3478 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
9251 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
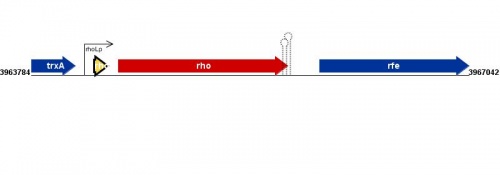 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Rho autoregulates transcription via the Rho attenuator in rhoL [14]
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:3964420..3964460
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for rho | |
|
microarray |
Summary of data for rho from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (3963867..3964103) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to rho Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10845 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0838 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10845 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000836 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012362 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
RHO |
From SHIGELLACYC |
|
E. coli O157 |
RHO |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
PF00006 ATP synthase alpha/beta family, nucleotide-binding domain |
||
|
EcoCyc:EG10845 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10845 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000836 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0838 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012362 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ Das, A et al. (1976) Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc. Natl. Acad. Sci. U.S.A. 73 1959-63 PubMed
- ↑ Opperman, T et al. (1995) The ts15 mutation of Escherichia coli alters the sequence of the C-terminal nine residues of Rho protein. Gene 152 133-4 PubMed
- ↑ Stewart, V & Yanofsky, C (1985) Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coli K-12. J. Bacteriol. 164 731-40 PubMed
- ↑ 5.0 5.1 Morse, DE & Guertin, M (1972) Amber suA mutations which relieve polarity. J. Mol. Biol. 63 605-8 PubMed
- ↑ 6.0 6.1 Ratner, D (1976) Evidence that mutations in the suA polarity suppressing gene directly affect termination factor rho. Nature 259 151-3 PubMed
- ↑ 7.0 7.1 Harinarayanan, R & Gowrishankar, J (2003) Host factor titration by chromosomal R-loops as a mechanism for runaway plasmid replication in transcription termination-defective mutants of Escherichia coli. J. Mol. Biol. 332 31-46 PubMed
- ↑ Yamamoto, N et al. (2009) Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 5 335 PubMed
- ↑ 9.0 9.1 CGSC: The Coli Genetics Stock Center
- ↑ 10.0 10.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ 11.00 11.01 11.02 11.03 11.04 11.05 11.06 11.07 11.08 11.09 11.10 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 12.0 12.1 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ Matsumoto, Y et al. (1986) Autogenous regulation of the gene for transcription termination factor rho in Escherichia coli: localization and function of its attenuators. J. Bacteriol. 166 945-58 PubMed
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories


