priA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
priA |
|---|---|
| Gene Synonym(s) |
ECK3927, b3935, JW3906, srgA[1], srgA |
| Product Desc. |
Primosome factor Y, also called protein n'; ATP-dependent DNA helicase activity required for recA-dependent stable DNA replication mode; also involved in double-strand break repair[4] |
| Product Synonyms(s) |
Primosome factor n' (replication factor Y)[1], B3935[2][1], SrgA[2][1], replication factor Y[2][1], PriA[2][1] , ECK3927, JW3906, srgA, b3935 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
priA |
|---|---|
| Mnemonic |
Primosome |
| Synonyms |
ECK3927, b3935, JW3906, srgA[1], srgA |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
88.86 minutes |
MG1655: 4124833..4122635 |
||
|
NC_012967: 4106293..4104095 |
||||
|
NC_012759: 4012304..4014502 |
||||
|
W3110 |
|
W3110: 3509871..3512069 |
||
|
DH10B: 4224530..4222332 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
4122635 |
Edman degradation |
PMID:2162049 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
priA(del) (Keio:JW3906) |
deletion |
deletion |
PMID:16738554 |
||||
|
priA1::KanR |
|
PMID:1938875 PMID:10713144 PMID:8631700 |
|||||
|
ΔpriA722::kan |
PMID:16738554 |
||||||
|
priA2::kan |
inserted kan gene fragment |
|
PMID:8631700 |
||||
|
priA2::kan ftsKC |
Growth Phenotype |
synthetically lethal |
PMID:11459954 |
||||
|
priA in MDS42 |
deletion |
Sensitivity to |
increases sensitivity to Bicyclomycin |
PMID:21183718 |
fig. 2 | ||
|
recF4101 priA2:kan |
Growth Phenotype |
Double mutation decreases cell viability from wildtype, table 1. |
PMID:8820655 |
experimental strain: JC18991 |
|||
|
recF4115 priA2:kan |
Growth Phenotype |
Double mutation decreases cell viability, table 1. |
PMID:8820655 |
experimental strain: JC18996 |
|||
|
recF143 priA2::kan |
Growth Phenotype |
Double mutation causes decreased cell viability, table 1. |
PMID:8820655 |
JC18989 |
|||
|
priA2::kan |
Sensitivity to |
Mutations shows increase in UV-inducible SOS expression, fig 1A. |
PMID:8820655 |
||||
|
priA::kanr |
Cell Shape |
Inactivation of priA showed huge multinucleoid cell filaments, Fig 4B. |
PMID:1938875 |
||||
|
priA::kanr |
Growth Phenotype |
Inactivation causes slow growth rate, fig 3. |
PMID:1938875 |
| |||
| edit table |
<protect></protect>
Notes
spa-47 (dnaC) suppresses the recombination and DSB repair defects seen in the priA1::kan mutant [6]
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW3906 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCCCCGTTGCCCACGTTGCCTT Primer 2:CCACCCTCAATCGGATCAACATC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 5% [7] | ||
|
Linked marker |
est. P1 cotransduction: 47% [7] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10763 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10763 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000754 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0756 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012860 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
PriA |
|---|---|
| Synonyms |
Primosome factor n' (replication factor Y)[1], B3935[2][1], SrgA[2][1], replication factor Y[2][1], PriA[2][1] , ECK3927, JW3906, srgA, b3935 |
| Product description |
Primosome factor Y, also called protein n'; ATP-dependent DNA helicase activity required for recA-dependent stable DNA replication mode; also involved in double-strand break repair[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003676 |
nucleic acid binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001650 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003676 |
nucleic acid binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011545 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005259 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0238 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004386 |
helicase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001650 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004386 |
helicase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0347 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001650 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011545 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005658 |
alpha DNA polymerase:primase complex |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0639 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006260 |
DNA replication |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005259 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006260 |
DNA replication |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0235 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006269 |
DNA replication, synthesis of RNA primer |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0639 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008026 |
ATP-dependent helicase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011545 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008270 |
zinc ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0862 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006261 |
DNA-dependent DNA replication |
PMID:10097074 |
IDA: Inferred from Direct Assay |
P |
required for D-loop dependent DNA replication |
complete | ||
|
GO:0016787 |
hydrolase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0378 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046872 |
metal ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0479 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006310 |
DNA recombination |
PMID:8631700 |
IMP: Inferred from Mutant Phenotype |
P |
defective in transductional and conjugative recombination |
complete | ||
|
GO:0010332 |
response to gamma radiation |
PMID:8631700 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0006302 |
double-strand break repair |
PMID:8631700 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0046677 |
response to antibiotic |
PMID:8631700 |
IMP: Inferred from Mutant Phenotype |
P |
hypersensitive to mitomycin C |
complete | ||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of primosome |
could be indirect |
||
|
Protein |
rplB |
PMID:16606699 |
Experiment(s):EBI-1147108 | |
|
Protein |
rpsC |
PMID:16606699 |
Experiment(s):EBI-1147108 | |
|
Protein |
galF |
PMID:16606699 |
Experiment(s):EBI-1147108 | |
|
Protein |
rhsA |
PMID:16606699 |
Experiment(s):EBI-1147108 | |
|
Protein |
rplQ |
PMID:16606699 |
Experiment(s):EBI-1147108 | |
|
Protein |
tufA |
PMID:15690043 |
Experiment(s):EBI-884556 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MPVAHVALPV PLPRTFDYLL PEGMTVKAGC RVRVPFGKQQ ERIGIVVSVS DASELPLNEL KAVVEVLDSE PVFTHSVWRL LLWAADYYHH PIGDVLFHAL PILLRQGRPA ANAPMWYWFA TEQGQAVDLN SLKRSPKQQQ ALAALRQGKI WRDQVATLEF NDAALQALRK KGLCDLASET PEFSDWRTNY AVSGERLRLN TEQATAVGAI HSAADTFSAW LLAGVTGSGK TEVYLSVLEN VLAQGKQALV MVPEIGLTPQ TIARFRERFN APVEVLHSGL NDSERLSAWL KAKNGEAAIV IGTRSALFTP FKNLGVIVID EEHDSSYKQQ EGWRYHARDL AVYRAHSEQI PIILGSATPA LETLCNVQQK KYRLLRLTRR AGNARPAIQH VLDLKGQKVQ AGLAPALITR MRQHLQADNQ VILFLNRRGF APALLCHDCG WIAECPRCDH YYTLHQAQHH LRCHHCDSQR PVPRQCPSCG STHLVPVGLG TEQLEQTLAP LFPGVPISRI DRDTTSRKGA LEQQLAEVHR GGARILIGTQ MLAKGHHFPD VTLVALLDVD GALFSADFRS AERFAQLYTQ VAGRAGRAGK QGEVVLQTHH PEHPLLQTLL YKGYDAFAEQ ALAERRMMQL PPWTSHVIVR AEDHNNQHAP LFLQQLRNLI LSSPLADEKL WVLGPVPALA PKRGGRWRWQ ILLQHPSRVR LQHIINGTLA LINTIPDSRK VKWVLDVDPI EG |
| Length |
732 |
| Mol. Wt |
81.654 kDa |
| pI |
9.1 (calculated) |
| Extinction coefficient |
104,850 - 106,225 (calc based on 15 Y, 15 W, and 11 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0012860 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10763 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10763 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000754 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0756 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
105 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
59 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
76 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
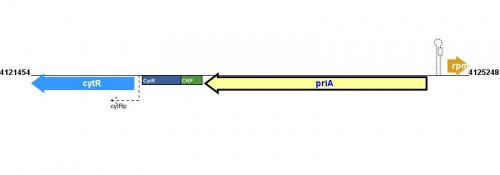 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:4124813..4124853
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for priA | |
|
microarray |
Summary of data for priA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (4124313..4124659) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to priA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10763 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0756 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10763 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000754 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012860 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Shigella flexneri |
PRIA |
From SHIGELLACYC |
|
E. coli O157 |
PRIA |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10763 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10763 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000754 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0756 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012860 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ Kogoma, T et al. (1996) The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J. Bacteriol. 178 1258-64 PubMed
- ↑ 7.0 7.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories


