pnp:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
pnp |
|---|---|
| Gene Synonym(s) |
ECK3152, b3164, JW5851, bfl[1], bfl |
| Product Desc. |
Component of polynucleotide phosphorylase[3]; degradosome[3] Polynucleotide phosphorylase; exoribonuclease; PNPase component of RNA degradosome; cold shock protein required for growth at low temperatures[4] |
| Product Synonyms(s) |
polynucleotide phosphorylase/polyadenylase[1], B3164[2][1], Bfl[2][1], Pnp[2][1], polynucleotide phosphorylase[2][1] , bfl, ECK3152, JW5851, b3164 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression |
transcription unit(s): rpsO-pnp[2], pnp[2], metY-yhbC-nusA-infB-rbfA-truB-rpsO-pnp[2] |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Implicated in degadation of sRNAs[5]
Cold induction occurs by reversal of post-translational autoregulation. The pnp mRNA leader is cleaved by RNase III, allowing PNPase to repress its own translation. Mutation of Gly454 impairs autogenous regulation without affecting catalytic activity. Mutants have elevated OM vesiculation (McBroom, 2006). Pnp can add long polynucleotide tails to mRNAs terminated by Rho factor (Mohanty, 2006). The overexpression of the Pnp S1 domain suppresses the cold growth defect of quadruple cspABEG mutant strain BX04 (Xia, 2001).[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
pnp |
|---|---|
| Mnemonic |
Polynucleotide phosphorylase |
| Synonyms |
ECK3152, b3164, JW5851, bfl[1], bfl |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
71.28 minutes |
MG1655: 3309259..3307055 |
||
|
NC_012967: 3246471..3244267 |
||||
|
NC_012759: 3194203..3196338 |
||||
|
W3110 |
|
W3110: 3311092..3308888 |
||
|
DH10B: 3407004..3404800 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
3307055 |
Edman degradation |
PMID:9298646 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
Δpnp (Keio:JW5851) |
deletion |
deletion |
PMID:16738554 |
||||
|
pnp::Tn5KAN-I-SceI (FB21050) |
Insertion at nt 556 in Plus orientation |
PMID:15262929 |
does not contain pKD46 | ||||
|
pnp::Tn5KAN-I-SceI (FB21051) |
Insertion at nt 1758 in Minus orientation |
PMID:15262929 |
contains pKD46 | ||||
|
pnp-13 |
|||||||
|
pnp-7 |
|||||||
|
pnp-27 |
| ||||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW5851 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCCGCAGAAGATCGGGTATTAA Primer 2:CCtTCGCCCTGTTCAGCAGCCGG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: % [7] | ||
|
Linked marker |
est. P1 cotransduction: 40% [7] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10743 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10743 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000734 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0736 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010397 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
Pnp |
|---|---|
| Synonyms |
polynucleotide phosphorylase/polyadenylase[1], B3164[2][1], Bfl[2][1], Pnp[2][1], polynucleotide phosphorylase[2][1] , bfl, ECK3152, JW5851, b3164 |
| Product description |
Component of polynucleotide phosphorylase[3]; degradosome[3] Polynucleotide phosphorylase; exoribonuclease; PNPase component of RNA degradosome; cold shock protein required for growth at low temperatures[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000175 |
3'-5'-exoribonuclease activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001247 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000175 |
3'-5'-exoribonuclease activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR015847 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000175 |
3'-5'-exoribonuclease activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR015848 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003723 |
RNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001247 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003723 |
RNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003029 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003723 |
RNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004087 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003723 |
RNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004088 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003723 |
RNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012162 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003723 |
RNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR015847 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003723 |
RNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR015848 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003723 |
RNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR018111 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003723 |
RNA binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0694 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004654 |
polyribonucleotide nucleotidyltransferase activity |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_01595 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004654 |
polyribonucleotide nucleotidyltransferase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012162 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004654 |
polyribonucleotide nucleotidyltransferase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:2.7.7.8 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005739 |
mitochondrion |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012162 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005829 |
cytosol |
PMID:16858726 |
IDA: Inferred from Direct Assay |
C |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0006396 |
RNA processing |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001247 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006396 |
RNA processing |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR015847 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006396 |
RNA processing |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR015848 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006402 |
mRNA catabolic process |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_01595 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006402 |
mRNA catabolic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012162 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006950 |
response to stress |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0346 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
PMID:16858726 |
IDA: Inferred from Direct Assay |
C |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0016740 |
transferase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0808 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016779 |
nucleotidyltransferase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0548 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of polynucleotide phosphorylase |
could be indirect |
||
|
Protein |
metK |
PMID:15690043 |
Experiment(s):EBI-883913 | |
|
Protein |
recA |
PMID:15690043 |
Experiment(s):EBI-883913 | |
|
Protein |
rplB |
PMID:15690043 |
Experiment(s):EBI-883913 | |
|
Protein |
rplD |
PMID:15690043 |
Experiment(s):EBI-883913, EBI-891089 | |
|
Protein |
rplF |
PMID:15690043 |
Experiment(s):EBI-883913 | |
|
Protein |
rplI |
PMID:15690043 |
Experiment(s):EBI-883913, EBI-891089 | |
|
Protein |
rplM |
PMID:15690043 |
Experiment(s):EBI-883913, EBI-891089 | |
|
Protein |
rpoA |
PMID:15690043 |
Experiment(s):EBI-883913 | |
|
Protein |
rpsC |
PMID:15690043 |
Experiment(s):EBI-883913 | |
|
Protein |
rpsD |
PMID:15690043 |
Experiment(s):EBI-883913 | |
|
Protein |
rpsE |
PMID:15690043 |
Experiment(s):EBI-883913, EBI-891089 | |
|
Protein |
rpsG |
PMID:15690043 |
Experiment(s):EBI-883913, EBI-891089 | |
|
Protein |
rpsM |
PMID:15690043 |
Experiment(s):EBI-883913, EBI-891089 | |
|
Protein |
srmB |
PMID:15690043 |
Experiment(s):EBI-883913 | |
|
Protein |
rplE |
PMID:15690043 |
Experiment(s):EBI-883913 | |
|
Protein |
pgk |
PMID:15690043 |
Experiment(s):EBI-891089, EBI-890008 | |
|
Protein |
rplA |
PMID:15690043 |
Experiment(s):EBI-891089 | |
|
Protein |
rplO |
PMID:15690043 |
Experiment(s):EBI-891089 | |
|
Protein |
rplQ |
PMID:15690043 |
Experiment(s):EBI-891089 | |
|
Protein |
rplU |
PMID:15690043 |
Experiment(s):EBI-891089 | |
|
Protein |
rplV |
PMID:15690043 |
Experiment(s):EBI-891089 | |
|
Protein |
rplX |
PMID:15690043 |
Experiment(s):EBI-891089 | |
|
Protein |
rpsO |
PMID:15690043 |
Experiment(s):EBI-891089 | |
|
Protein |
rpsR |
PMID:15690043 |
Experiment(s):EBI-891089 | |
|
Protein |
rpsS |
PMID:15690043 |
Experiment(s):EBI-891089 | |
|
Protein |
rpsT |
PMID:15690043 |
Experiment(s):EBI-891089 | |
|
Protein |
hupA |
PMID:15690043 |
Experiment(s):EBI-891089 | |
|
Protein |
rpsS |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplE |
PMID:19402753 |
MALDI(Z-score):28.336587 | |
|
Protein |
rpsR |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
eno |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):39.324472 | |
|
Protein |
rplO |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):3.209278 | |
|
Protein |
rpsO |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
hupA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rpsM |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):20.758343 | |
|
Protein |
rpsT |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplQ |
PMID:19402753 |
LCMS(ID Probability):99.4 | |
|
Protein |
rplI |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):18.361833 | |
|
Protein |
rplU |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplX |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rpsC |
PMID:19402753 |
MALDI(Z-score):35.080787 | |
|
Protein |
srmB |
PMID:19402753 |
MALDI(Z-score):26.241773 | |
|
Protein |
rplM |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):8.655067 | |
|
Protein |
rpsD |
PMID:19402753 |
LCMS(ID Probability):95.3 MALDI(Z-score):19.991630 | |
|
Protein |
rplB |
PMID:19402753 |
LCMS(ID Probability):94.5 MALDI(Z-score):35.593951 | |
|
Protein |
rplA |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):16.114612 | |
|
Protein |
rplD |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):16.007735 | |
|
Protein |
rhlB |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):37.285496 | |
|
Protein |
deaD |
PMID:19402753 |
MALDI(Z-score):18.761801 | |
|
Protein |
rluB |
PMID:19402753 |
MALDI(Z-score):33.707783 | |
|
Protein |
Subunits of degradosome |
could be indirect |
| |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MRRRSGINTS AVRYCLRKRK DITLLNPIVR KFQYGQHTVT LETGMMARQA TAAVMVSMDD TAVFVTVVGQ KKAKPGQDFF PLTVNYQERT YAAGRIPGSF FRREGRPSEG ETLIARLIDR PIRPLFPEGF VNEVQVIATV VSVNPQVNPD IVAMIGASAA LSLSGIPFNG PIGAARVGYI NDQYVLNPTQ DELKESKLDL VVAGTEAAVL MVESEAQLLS EDQMLGAVVF GHEQQQVVIQ NINELVKEAG KPRWDWQPEP VNEALNARVA ALAEARLSDA YRITDKQERY AQVDVIKSET IATLLAEDET LDENELGEIL HAIEKNVVRS RVLAGEPRID GREKDMIRGL DVRTGVLPRT HGSALFTRGE TQALVTATLG TARDAQVLDE LMGERTDTFL FHYNFPPYSV GETGMVGSPK RREIGHGRLA KRGVLAVMPD MDKFPYTVRV VSEITESNGS SSMASVCGAS LALMDAGVPI KAAVAGIAMG LVKEGDNYVV LSDILGDEDH LGDMDFKVAG SRDGISALQM DIKIEGITKE IMQVALNQAK GARLHILGVM EQAINAPRGD ISEFAPRIHT IKINPDKIKD VIGKGGSVIR ALTEETGTTI EIEDDGTVKI AATDGEKAKH AIRRIEEITA EIEVGRVYTG KVTRIVDFGA FVAIGGGKEG LVHISQIADK RVEKVTDYLQ MGQEVPVKVL EVDRQGRIRL SIKEATEQSQ PAAAPEAPAA EQGE |
| Length |
734 |
| Mol. Wt |
79.845 kDa |
| pI |
5.3 (calculated) |
| Extinction coefficient |
31,860 - 32,110 (calc based on 14 Y, 2 W, and 2 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0010397 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10743 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10743 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000734 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0736 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
5.70E+03 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
938.13+/-5.62 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.3027+/-0.0129 |
Molecules/cell |
|
by FISH |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.423548094 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 EMG2 |
200 |
molecules/cell |
|
Quantitative protein sequencing |
Spot ID: M59 |
PMID: 9298646 |
|
Protein |
E. coli K-12 MG1655 |
11905 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
3545 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
7930 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
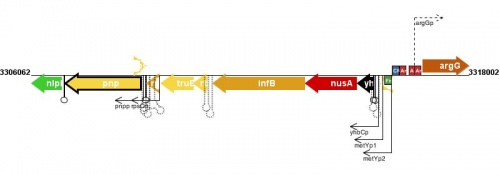 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:3309239..3309279
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for pnp | |
|
microarray |
Summary of data for pnp from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (3308790..3309137) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to pnp Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10743 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0736 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10743 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000734 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010397 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Drosophila melanogaster |
|
From Inparanoid:20070104 |
|
Drosophila pseudoobscura |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Macaca mulatta |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Tetraodon nigroviridis |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
PNP |
From SHIGELLACYC |
|
E. coli O157 |
PNP |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
PF03726 Polyribonucleotide nucleotidyltransferase, RNA binding domain |
||
|
EcoCyc:EG10743 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10743 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000734 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0736 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010397 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ Andrade, JM & Arraiano, CM (2008) PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA 14 543-51 PubMed
- ↑ 6.0 6.1 CGSC: The Coli Genetics Stock Center
- ↑ 7.0 7.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories
- Genes in OpenBioSystems with Promoter Fusions
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Apis mellifera
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Bos taurus
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Canis familiaris
- Genes with homologs in Drosophila melanogaster
- Genes with homologs in Drosophila pseudoobscura
- Genes with homologs in Gallus gallus
- Genes with homologs in Homo sapiens
- Genes with homologs in Macaca mulatta
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Oryza gramene
- Genes with homologs in Pan troglodytes
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Schizosaccharomyces pombe
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Tetraodon nigroviridis
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


