metH:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
metH |
|---|---|
| Gene Synonym(s) |
ECK4011, b4019, JW3979[1], JW3979 |
| Product Desc. |
Methionine synthase, cobalamin-dependent; 5-methyltetrahydrofolate-homocysteine methyltransferase; binds Zn(II)[2] |
| Product Synonyms(s) |
homocysteine-N5-methyltetrahydrofolate transmethylase, B12-dependent[1], B4019[3][1], MetH[3][1] , ECK4011, JW3979, b4019 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
metH |
|---|---|
| Mnemonic |
Methionine |
| Synonyms |
ECK4011, b4019, JW3979[1], JW3979 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
90.99 minutes |
MG1655: 4221851..4225534 |
||
|
NC_012967: 4202759..4206442 |
||||
|
NC_012759: 4111531..4115214 |
||||
|
W3110 |
|
W3110: 4227418..4231101 |
||
|
DH10B: 4321547..4325230 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
4221854 |
Edman degradation |
PMID:2668277 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔmetH (Keio:JW3979) |
deletion |
deletion |
PMID:16738554 |
||||
|
metHC310A,S |
C310A,S |
Loss of zinc binding. Loss of catalytic activity |
seeded from UniProt:P13009 | ||||
|
metHC311A,S |
C311A,S |
Loss of zinc binding. Loss of catalytic activity |
seeded from UniProt:P13009 | ||||
|
metHD757E |
D757E |
Decreases activity by about 70% |
seeded from UniProt:P13009 | ||||
|
metHD757N |
D757N |
Decreases activity by about 45% |
seeded from UniProt:P13009 | ||||
|
metHH759G |
H759G |
Loss of catalytic activity |
seeded from UniProt:P13009 | ||||
|
metHS810A |
S810A |
Decreases activity by about 40% |
seeded from UniProt:P13009 | ||||
|
metH156 |
PMID:4579870 |
||||||
|
ΔmetH786::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW3979 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAGCAGCAAAGTGGAACAACT Primer 2:CCGTCCGCGTCATACCCCAGATT | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 35% [5] | ||
|
Linked marker |
est. P1 cotransduction: 51% [5] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10587 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10587 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000580 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0582 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0013140 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
MetH |
|---|---|
| Synonyms |
homocysteine-N5-methyltetrahydrofolate transmethylase, B12-dependent[1], B4019[3][1], MetH[3][1] , ECK4011, JW3979, b4019 |
| Product description |
Methionine synthase, cobalamin-dependent; 5-methyltetrahydrofolate-homocysteine methyltransferase; binds Zn(II)[2] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0005737 |
cytoplasm |
C |
Seeded from Riley et al 2006 [1]. |
Missing: evidence, reference | ||||
|
GO:0004156 |
dihydropteroate synthase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000489 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005622 |
intracellular |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004223 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008168 |
methyltransferase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0489 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008270 |
zinc ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0862 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008652 |
cellular amino acid biosynthetic process |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0028 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008705 |
methionine synthase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003759 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008705 |
methionine synthase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004223 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008705 |
methionine synthase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011822 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008705 |
methionine synthase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:2.1.1.13 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
PMID:9250984 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0008898 |
homocysteine S-methyltransferase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003726 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
PMID:2668277 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0009086 |
methionine biosynthetic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003759 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008705 |
methionine synthase activity |
PMID:10978155 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0009086 |
methionine biosynthetic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004223 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008168 |
methyltransferase activity |
Taylor and Weissbach (1971)[6] |
IDA: Inferred from Direct Assay |
F |
Missing: reference | |||
|
GO:0009086 |
methionine biosynthetic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011822 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008898 |
homocysteine S-methyltransferase activity |
Taylor and Weissbach (1971)[6] |
IDA: Inferred from Direct Assay |
F |
Missing: reference | |||
|
GO:0009086 |
methionine biosynthetic process |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0486 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009257 |
10-formyltetrahydrofolate biosynthetic process |
Taylor and Weissbach (1971)[6] |
IDA: Inferred from Direct Assay |
P |
Missing: reference | |||
|
GO:0009396 |
folic acid and derivative biosynthetic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000489 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008705 |
methionine synthase activity |
Taylor and Weissbach (1971)[6] |
IDA: Inferred from Direct Assay |
F |
Missing: reference | |||
|
GO:0031419 |
cobalamin binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003759 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008652 |
cellular amino acid biosynthetic process |
Taylor and Weissbach (1971)[6] |
IDA: Inferred from Direct Assay |
P |
Missing: reference | |||
|
GO:0031419 |
cobalamin binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006158 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009086 |
methionine biosynthetic process |
Taylor and Weissbach (1971)[6] |
IDA: Inferred from Direct Assay |
P |
Missing: reference | |||
|
GO:0031419 |
cobalamin binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011822 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0031419 |
cobalamin binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0846 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0044237 |
cellular metabolic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011005 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046872 |
metal ion binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003759 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046872 |
metal ion binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006158 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046872 |
metal ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0479 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0050897 |
cobalt ion binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011822 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0050897 |
cobalt ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0170 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
gltB |
PMID:16606699 |
Experiment(s):EBI-1147304 | |
|
Protein |
dnaE |
PMID:16606699 |
Experiment(s):EBI-1147304 | |
|
Protein |
ilvC |
PMID:16606699 |
Experiment(s):EBI-1147304 | |
|
Protein |
bglX |
PMID:16606699 |
Experiment(s):EBI-1147304 | |
|
Small Molecule |
Cobalamin |
PMID:9201956 |
| |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MSSKVEQLRA QLNERILVLD GGMGTMIQSY RLNEADFRGE RFADWPCDLK GNNDLLVLSK PEVIAAIHNA YFEAGADIIE TNTFNSTTIA MADYQMESLS AEINFAAAKL ARACADEWTA RTPEKPRYVA GVLGPTNRTA SISPDVNDPA FRNITFDGLV AAYRESTKAL VEGGADLILI ETVFDTLNAK AAVFAVKTEF EALGVELPIM ISGTITDASG RTLSGQTTEA FYNSLRHAEA LTFGLNCALG PDELRQYVQE LSRIAECYVT AHPNAGLPNA FGEYDLDADT MAKQIREWAQ AGFLNIVGGC CGTTPQHIAA MSRAVEGLAP RKLPEIPVAC RLSGLEPLNI GEDSLFVNVG ERTNVTGSAK FKRLIKEEKY SEALDVARQQ VENGAQIIDI NMDEGMLDAE AAMVRFLNLI AGEPDIARVP IMIDSSKWDV IEKGLKCIQG KGIVNSISMK EGVDAFIHHA KLLRRYGAAV VVMAFDEQGQ ADTRARKIEI CRRAYKILTE EVGFPPEDII FDPNIFAVAT GIEEHNNYAQ DFIGACEDIK RELPHALISG GVSNVSFSFR GNDPVREAIH AVFLYYAIRN GMDMGIVNAG QLAIYDDLPA ELRDAVEDVI LNRRDDGTER LLELAEKYRG SKTDDTANAQ QAEWRSWEVN KRLEYSLVKG ITEFIEQDTE EARQQATRPI EVIEGPLMDG MNVVGDLFGE GKMFLPQVVK SARVMKQAVA YLEPFIEASK EQGKTNGKMV IATVKGDVHD IGKNIVGVVL QCNNYEIVDL GVMVPAEKIL RTAKEVNADL IGLSGLITPS LDEMVNVAKE MERQGFTIPL LIGGATTSKA HTAVKIEQNY SGPTVYVQNA SRTVGVVAAL LSDTQRDDFV ARTRKEYETV RIQHGRKKPR TPPVTLEAAR DNDFAFDWQA YTPPVAHRLG VQEVEASIET LRNYIDWTPF FMTWSLAGKY PRILEDEVVG VEAQRLFKDA NDMLDKLSAE KTLNPRGVVG LFPANRVGDD IEIYRDETRT HVINVSHHLR QQTEKTGFAN YCLADFVAPK LSGKADYIGA FAVTGGLEED ALADAFEAQH DDYNKIMVKA LADRLAEAFA EYLHERVRKV YWGYAPNENL SNEELIRENY QGIRPAPGYP ACPEHTEKAT IWELLEVEKH TGMKLTESFA MWPGASVSGW YFSHPDSKYY AVAQIQRDQV EDYARRKGMS VTEVERWLAP NLGYDAD |
| Length |
1,227 |
| Mol. Wt |
135.998 kDa |
| pI |
4.8 (calculated) |
| Extinction coefficient |
136,600 - 138,225 (calc based on 40 Y, 14 W, and 13 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0013140 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10587 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10587 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000580 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0582 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
1.24E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
E. coli K-12 MG1655 |
559 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
753 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
1780 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
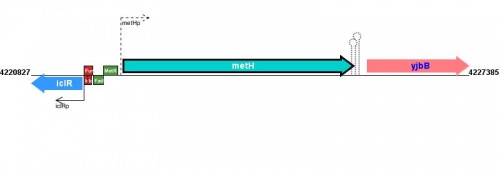 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:4221831..4221871
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for metH | |
|
microarray |
Summary of data for metH from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to metH Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10587 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0582 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10587 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000580 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0013140 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Danio rerio |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Macaca mulatta |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Tetraodon nigroviridis |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
METH |
From SHIGELLACYC |
|
E. coli O157 |
METH |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
PF02965 Vitamin B12 dependent methionine synthase, activation domain |
||
|
EcoCyc:EG10587 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10587 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000580 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0582 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0013140 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 3.0 3.1 3.2 3.3 3.4 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 CGSC: The Coli Genetics Stock Center
- ↑ 5.0 5.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 Taylor, R.T. and Weissbach, H. NS-methyltetrahydrofolate-homocysteine
(Vitamin B12) methyltransferase (Escherichia coli B).
Methods Enzymol. 17B (1971) 379-388.Taylor & Weissbach Cite error: Invalid
<ref>tag; name "Taylor" defined multiple times with different content Cite error: Invalid<ref>tag; name "Taylor" defined multiple times with different content Cite error: Invalid<ref>tag; name "Taylor" defined multiple times with different content Cite error: Invalid<ref>tag; name "Taylor" defined multiple times with different content Cite error: Invalid<ref>tag; name "Taylor" defined multiple times with different content
Categories
- Pages with reference errors
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Bos taurus
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Canis familiaris
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Danio rerio
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Gallus gallus
- Genes with homologs in Homo sapiens
- Genes with homologs in Macaca mulatta
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Pan troglodytes
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Tetraodon nigroviridis
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


