entF:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
entF |
|---|---|
| Gene Synonym(s) |
ECK0579, b0586, JW0578[1], JW0578 |
| Product Desc. |
Component of enterobactin synthase multienzyme complex[2][3] Enterochelin synthase, component F; multifunctional: holo-EntF(PCP), primed by EntD; seryl-AMP, seryl-S-EntF synthase; DHB-Ser-S-EntF intermediate formation; chain-releasing thioesterase (TE) domain is both a cyclotrimerizing lactone synthase and an elong[4] |
| Product Synonyms(s) |
enterobactin synthase multienzyme complex component, ATP-dependent[1], B0586 [2][1], holo-serine activating enzyme [2][1],apo-serine activating enzyme[2][1], ECK0579, JW0578, b0586 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression |
transcription unit(s): fes-ybdZ-entF-fepE[2] |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
entF |
|---|---|
| Mnemonic |
Enterochelin |
| Synonyms |
ECK0579, b0586, JW0578[1], JW0578 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
13.22 minutes |
MG1655: 613380..617261 |
||
|
NC_012967: 596865..600746 |
||||
|
NC_012759: 516140..520021 |
||||
|
W3110 |
|
W3110: 613380..617261 |
||
|
DH10B: 552712..556593 |
||||
|
DH10B: 665972..669853 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
613380 |
Edman degradation |
PMID:1826089 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔentF (Keio:JW0578) |
deletion |
deletion |
PMID:16738554 |
||||
|
entF::Tn5KAN-2 (FB20204) |
Insertion at nt 292 in Minus orientation |
PMID:15262929 |
does not contain pKD46 | ||||
|
entFS1138A |
S1138A |
No enterobactin synthetase activity. This mutant holo-entF is still able to adenylate serine and to autoacylate itself with serine |
seeded from UniProt:P11454 | ||||
|
ΔentF724::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW0578 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAGCCAGCATTTACCTTTGGT Primer 2:CCCCTGTTTAGCGTTGCGCGAAT | |
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 4% [6] | ||
|
Linked marker |
est. P1 cotransduction: 44% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10264 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10264 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000258 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0260 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0002021 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
EntF |
|---|---|
| Synonyms |
enterobactin synthase multienzyme complex component, ATP-dependent[1], B0586 [2][1], holo-serine activating enzyme [2][1],apo-serine activating enzyme[2][1], ECK0579, JW0578, b0586 |
| Product description |
Component of enterobactin synthase multienzyme complex[2][3] Enterochelin synthase, component F; multifunctional: holo-EntF(PCP), primed by EntD; seryl-AMP, seryl-S-EntF synthase; DHB-Ser-S-EntF intermediate formation; chain-releasing thioesterase (TE) domain is both a cyclotrimerizing lactone synthase and an elong[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000036 |
acyl carrier activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR009081 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003824 |
catalytic activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000873 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003824 |
catalytic activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0511 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005506 |
iron ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0408 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006810 |
transport |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0813 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006811 |
ion transport |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0406 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006826 |
iron ion transport |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0410 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008152 |
metabolic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000873 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009058 |
biosynthetic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001031 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009239 |
enterobactin biosynthetic process |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0259 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016740 |
transferase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0808 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016788 |
hydrolase activity, acting on ester bonds |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001031 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016874 |
ligase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR010071 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016874 |
ligase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0436 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0031177 |
phosphopantetheine binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006162 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0048037 |
cofactor binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006163 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of enterobactin synthase multienzyme complex |
could be indirect |
||
|
Protein |
ndh |
PMID:16606699 |
Experiment(s):EBI-1137240 | |
|
Protein |
carA |
PMID:16606699 |
Experiment(s):EBI-1137240 | |
|
Protein |
tatE |
PMID:16606699 |
Experiment(s):EBI-1137240 | |
|
Protein |
groL |
PMID:16606699 |
Experiment(s):EBI-1137240 | |
|
Protein |
ycgX |
PMID:16606699 |
Experiment(s):EBI-1137240 | |
| edit table |
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
plasma membrane |
From EcoCyc[3] |
| ||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MSQHLPLVAA QPGIWMAEKL SELPSAWSVA HYVELTGEVD SPLLARAVVA GLAQADTLRM RFTEDNGEVW QWVDDALTFE LPEIIDLRTN IDPHGTAQAL MQADLQQDLR VDSGKPLVFH QLIQVADNRW YWYQRYHHLL VDGFSFPAIT RQIANIYCTW LRGEPTPASP FTPFADVVEE YQQYRESEAW QRDAAFWAEQ RRQLPPPASL SPAPLPGRSA SADILRLKLE FTDGEFRQLA TQLSGVQRTD LALALAALWL GRLCNRMDYA AGFIFMRRLG SAALTATGPV LNVLPLGIHI AAQETLPELA TRLAAQLKKM RRHQRYDAEQ IVRDSGRAAG DEPLFGPVLN IKVFDYQLDI PDVQAQTHTL ATGPVNDLEL ALFPDVHGDL SIEILANKQR YDEPTLIQHA ERLKMLIAQF AADPALLCGD VDIMLPGEYA QLAQLNATQV EIPETTLSAL VAEQAAKTPD APALADARYL FSYREMREQV VALANLLRER GVKPGDSVAV ALPRSVFLTL ALHAIVEAGA AWLPLDTGYP DDRLKMMLED ARPSLLITTD DQLPRFSDVP NLTSLCYNAP LTPQGSAPLQ LSQPHHTAYI IFTSGSTGRP KGVMVGQTAI VNRLLWMQNH YPLTGEDVVA QKTPCSFDVS VWEFFWPFIA GAKLVMAEPE AHRDPLAMQQ FFAEYGVTTT HFVPSMLAAF VASLTPQTAR QSCATLKQVF CSGEALPADL CREWQQLTGA PLHNLYGPTE AAVDVSWYPA FGEELAQVRG SSVPIGYPVW NTGLRILDAM MHPVPPGVAG DLYLTGIQLA QGYLGRPDLT ASRFIADPFA PGERMYRTGD VARWLDNGAV EYLGRSDDQL KIRGQRIELG EIDRVMQALP DVEQAVTHAC VINQAAATGG DARQLVGYLV SQSGLPLDTS ALQAQLRETL PPHMVPVVLL QLPQLPLSAN GKLDRKALPL PELKAQAPGR APKAGSETII AAAFSSLLGC DVQDADADFF ALGGHSLLAM KLAAQLSRQV ARQVTPGQVM VASTVAKLAT IIDAEEDSTR RMGFETILPL REGNGPTLFC FHPASGFAWQ FSVLSRYLDP QWSIIGIQSP RPNGPMQTAA NLDEVCEAHL ATLLEQQPHG PYYLLGYSLG GTLAQGIAAR LRARGEQVAF LGLLDTWPPE TQNWQEKEAN GLDPEVLAEI NREREAFLAA QQGSTSTELF TTIEGNYADA VRLLTTAHSV PFDGKATLFV AERTLQEGMS PERAWSPWIA ELDIYRQDCA HVDIISPGTF EKIGPIIRAT LNR |
| Length |
1,293 |
| Mol. Wt |
141.993 kDa |
| pI |
4.9 (calculated) |
| Extinction coefficient |
181,170 - 182,795 (calc based on 33 Y, 24 W, and 13 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0002021 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10264 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10264 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000258 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0260 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
4.07E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
32.275+/-0.314 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.006570472 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
24 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
24 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
4a |
molecules/cell/generation |
|
Ribosome Profiling |
Low confidence in the sequencing data set. |
PMID: 24766808 |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
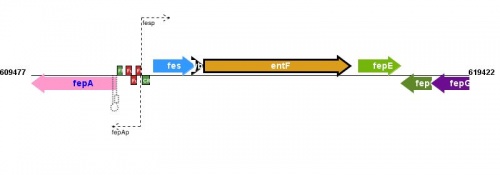 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:613360..613400
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for entF | |
|
microarray |
Summary of data for entF from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (613082..613445) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to entF Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10264 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0260 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10264 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000258 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0002021 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Drosophila melanogaster |
|
From Inparanoid:20070104 |
|
Drosophila pseudoobscura |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
ENTF |
From SHIGELLACYC |
|
E. coli O157 |
ENTF |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10264 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10264 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000258 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0260 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0002021 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 3.4 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories
- Genes in OpenBioSystems with Promoter Fusions
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Apis mellifera
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Drosophila melanogaster
- Genes with homologs in Drosophila pseudoobscura
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


