uvrA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
uvrA |
|---|---|
| Gene Synonym(s) |
ECK4050, b4058, JW4019, dar, dinE[1] |
| Product Desc. |
excision nuclease subunit A[2][3]; Component of UvrABC Nucleotide Excision Repair Complex[2][3] Excision nuclease subunit A; repair of UV damage to DNA; LexA regulon; binds Zn(II)[4] |
| Product Synonyms(s) |
ATPase and DNA damage recognition protein of nucleotide excision repair excinuclease UvrABC[1], B4058[2][1], Dar[2][1], DinE[2][1], UvrA[2][1] , dar, ECK4050, JW4019, b4058 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
uvrA |
|---|---|
| Mnemonic |
Ultraviolet resistant |
| Synonyms |
ECK4050, b4058, JW4019, dar, dinE[1] |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
92.01 minutes, 92.01 minutes |
MG1655: 4271894..4269072 |
||
|
NC_012967: 4252803..4249981 |
||||
|
NC_012759: 4207917..4210739 |
||||
|
W3110 |
|
W3110: 4277461..4274639 |
||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
4269072 |
Edman degradation |
PMID:3007478 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
Strain DH10B |
V332L |
Nonsynonomous mutation |
PMID:18245285 |
This is a SNP in E. coli K-12 Strain DH10B | |||
|
uvrA(del) (Keio:JW4019) |
deletion |
deletion |
PMID:16738554 |
||||
|
uvrAC253A,H,S |
C253A,H,S |
Reduced activity |
seeded from UniProt:P0A698 | ||||
|
uvrA6 |
Sensitivity to |
Mutation increases sensitivity to UV-radiation |
PMID:5335128 |
Figure 5 Strain AB1157 | |||
|
uvrA157 |
|||||||
|
uvrA103 |
|||||||
|
uvrA155 |
|||||||
|
uvrA277::Tn10 |
|||||||
|
uvrA37 |
Sensitivity to |
Mutation increases sensitivity to UV-radiation |
PMID:5335128 |
Figure 5 Strain Hfr J2 | |||
|
uvrA14 |
|||||||
|
uvrA18 |
|||||||
|
uvrA19 |
|||||||
|
uvrA281 |
|||||||
|
uvrA154 |
|||||||
|
uvrA0 |
|||||||
|
uvrA215::Mud(Ap,lac) |
PMID:6771759 PMID:6780917 |
||||||
|
uvrA753(del)::kan |
PMID:16738554 |
||||||
|
uvrA(del)::Tn10 |
deletion |
deletion |
Sensitivity to |
Deletion mutants are more sensitive to DNA damaging agent Mitomycin C, fig 1 & 2. |
PMID:17040104 |
Experimental strain:GY5564 |
|
|
uvrA::Tn10 uvrC34 |
Sensitivity to |
Double mutation increased sensitivity to mitomycin C but at a slower rate than single deletions. |
PMID:17040104 |
Strain: |
See Figure 1. | ||
|
uvrA(del) |
deletion |
deletion |
Sensitivity to |
Deletion mutation causes hypersensitive to UV when irradiated and incubated in non-photoreactivating conditions, fig 2. |
PMID:16970815 |
figure 2 | |
|
uvrA K689A |
Lysine to Alanine |
change in binding affinity |
Mutation caused a decrease in binding affinity to DNA, table 1. |
PMID:18248777 |
|||
|
uvrA K680E |
Lysine to Glutamic Acid |
Change in binding affinity |
Mutation caused a decrease in binding affinity to DNA, table 1. |
PMID:18248777 |
|||
|
uvrA R691A |
Arginine to Alanine |
Change in DNA binding affinity |
Mutation caused a decrease in binding affinity to DNA, table 1. |
PMID:18248777 |
|||
|
uvrA R691D |
Arginine to Aspartic Acid |
Change in binding affinity |
Mutation caused a decrease in binding affinity to DNA, table 1. |
PMID:18248777 |
|||
|
uvrA K680A R691A |
Lysine to Alanine & Arginine to Alanine |
Change in binding affinity |
Double mutation caused a decrease in binding affinity to DNA more than single point mutations, table 1. |
PMID:18248777 |
|||
|
uvrA::Tn10 uvrC34 |
DNA repair |
Double mutation was able to recover high molecular weight DNA, though at a much slower rate than wild type. |
PMID:17040104 |
Figure 4 | |||
|
uvrAB K45A |
Lysine to alanine |
change in enzyme activity |
ATP hydrolysis of uvrB by ATPase is inhibited (but not of uvrA in the complex). Table 1. |
PMID:17630776 |
|||
|
ZnG uvrA |
11 amino acid change, figure 1. |
DNA binding |
Mutant shows decreased binding affinity to damaged DNA, table 1. |
PMID:16829526 |
|||
|
uvrA G502D V508D |
Glycine to Aspartic Acid & Valine to Aspartic Acid |
Sensitivity to |
Mutation increases sensitivity to UV-radiation by causing the loss of uvrA's binding specificity between damaged and undamaged DNA. Figure 7. |
PMID:8444906 |
Figure 7 Parent Strain: MH1-∆A | ||
|
uvrA G275A |
Glycine to Alanine |
Resistant to |
Mutation creates "hyper" UV resistance greater than the UV resistance of wild type, figure 4. |
PMID:8444906 |
Figure 4 | ||
|
uvrA C763F |
Cysteine to Phenylalanine |
Change in DNA binding |
Mutation causes loss of single-stranded DNA-binding activity. |
PMID:8444906 |
No data shown Parent Strain: MH1-A(del) | ||
|
uvrA C740F H754Y C742Y/K750I |
Cysteine to Phenylalanine & Histidine to Tyrosine & Cysteine to Tyrosine/Lysine to Isoleucine (double mutant) |
Sensitivity to |
Mutation increased UV sensitivity by making the mutant proteins excision repair defective |
PMID:8444906 |
No data shown in source. Parent Strain: MH1-A(del) | ||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW4019 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCGATAAGATCGAAGTTCGGGG Primer 2:CCaAGCATCGGCTTAAGGAAGCG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 33% [6] | ||
|
Linked marker |
est. P1 cotransduction: 22% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11061 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11061 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001050 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB1054 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0013290 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
UvrA |
|---|---|
| Synonyms |
ATPase and DNA damage recognition protein of nucleotide excision repair excinuclease UvrABC[1], B4058[2][1], Dar[2][1], DinE[2][1], UvrA[2][1] , dar, ECK4050, JW4019, b4058 |
| Product description |
excision nuclease subunit A[2][3]; Component of UvrABC Nucleotide Excision Repair Complex[2][3] Excision nuclease subunit A; repair of UV damage to DNA; LexA regulon; binds Zn(II)[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00205 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0238 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004518 |
nuclease activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0267 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00205 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003439 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR017871 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00205 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0963 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0086 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006281 |
DNA repair |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00205 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006281 |
DNA repair |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0228 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006281 |
DNA repair |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0234 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006289 |
nucleotide-excision repair |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00205 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006289 |
nucleotide-excision repair |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004602 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006974 |
response to DNA damage stimulus |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0227 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008270 |
zinc ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0862 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009380 |
excinuclease repair complex |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004602 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009381 |
excinuclease ABC activity |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00205 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009381 |
excinuclease ABC activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004602 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009432 |
SOS response |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00205 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009432 |
SOS response |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0742 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016887 |
ATPase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003439 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016887 |
ATPase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR017871 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046872 |
metal ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0479 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of UvrABC Nucleotide Excision Repair Complex |
could be indirect |
||
|
Protein |
narG |
PMID:16606699 |
Experiment(s):EBI-1147356 | |
|
Protein |
narZ |
PMID:16606699 |
Experiment(s):EBI-1147356 | |
|
Protein |
dnaK |
PMID:16606699 |
Experiment(s):EBI-1147356 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MDKIEVRGAR THNLKNINLV IPRDKLIVVT GLSGSGKSSL AFDTLYAEGQ RRYVESLSAY ARQFLSLMEK PDVDHIEGLS PAISIEQKST SHNPRSTVGT ITEIHDYLRL LFARVGEPRC PDHDVPLAAQ TVSQMVDNVL SQPEGKRLML LAPIIKERKG EHTKTLENLA SQGYIRARID GEVCDLSDPP KLELQKKHTI EVVVDRFKVR DDLTQRLAES FETALELSGG TAVVADMDDP KAEELLFSAN FACPICGYSM RELEPRLFSF NNPAGACPTC DGLGVQQYFD PDRVIQNPEL SLAGGAIRGW DRRNFYYFQM LKSLADHYKF DVEAPWGSLS ANVHKVVLYG SGKENIEFKY MNDRGDTSIR RHPFEGVLHN MERRYKETES SAVREELAKF ISNRPCASCE GTRLRREARH VYVENTPLPA ISDMSIGHAM EFFNNLKLAG QRAKIAEKIL KEIGDRLKFL VNVGLNYLTL SRSAETLSGG EAQRIRLASQ IGAGLVGVMY VLDEPSIGLH QRDNERLLGT LIHLRDLGNT VIVVEHDEDA IRAADHVIDI GPGAGVHGGE VVAEGPLEAI MAVPESLTGQ YMSGKRKIEV PKKRVPANPE KVLKLTGARG NNLKDVTLTL PVGLFTCITG VSGSGKSTLI NDTLFPIAQR QLNGATIAEP APYRDIQGLE HFDKVIDIDQ SPIGRTPRSN PATYTGVFTP VRELFAGVPE SRARGYTPGR FSFNVRGGRC EACQGDGVIK VEMHFLPDIY VPCDQCKGKR YNRETLEIKY KGKTIHEVLD MTIEEAREFF DAVPALARKL QTLMDVGLTY IRLGQSATTL SGGEAQRVKL ARELSKRGTG QTLYILDEPT TGLHFADIQQ LLDVLHKLRD QGNTIVVIEH NLDVIKTADW IVDLGPEGGS GGGEILVSGT PETVAECEAS HTARFLKPML |
| Length |
940 |
| Mol. Wt |
103.869 kDa |
| pI |
6.6 (calculated) |
| Extinction coefficient |
53,750 - 55,500 (calc based on 25 Y, 3 W, and 14 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0013290 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG11061 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11061 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120001050 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1054 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
3.22E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
4.904+/-0.038 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.041814316 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
621 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
206 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
308 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
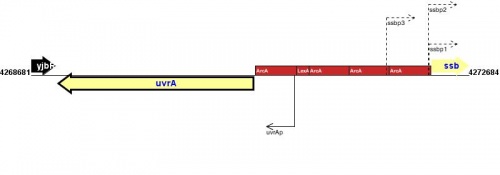 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:4271874..4271914
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for uvrA | |
|
microarray |
Summary of data for uvrA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (4271368..4271753) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to uvrA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11061 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1054 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11061 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001050 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0013290 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
UVRA |
From SHIGELLACYC |
|
E. coli O157 |
UVRA |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11061 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11061 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001050 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1054 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0013290 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories


