dnaX:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
dnaX |
|---|---|
| Gene Synonym(s) |
ECK0464, b0470, JW0459, dnaZ[1], dnaZ |
| Product Desc. |
|
| Product Synonyms(s) |
DNA polymerase III/DNA elongation factor III, tau and gamma subunits[1], DnaZ[2][1], B0470[2][1] , dnaZ, ECK0464, JW0459, b0470 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
The tau chain is the full length protein. The gamma chain is formed by programmed ribosomal frameshifting at codon 431 to a premature stop codon.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name | |
|---|---|
| Mnemonic | |
| Synonyms |
|
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
10.59 minutes |
MG1655: 491316..493247 |
||
|
NC_012967: 464159..466090 |
||||
|
NC_012759: 394075..396006 |
||||
|
W3110 |
|
W3110: 491316..493247 |
||
|
DH10B: 430647..432578 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
491319 |
Edman degradation |
PMID:3283125 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
dnaX36 |
E601K |
Growth Phenotype |
ts growth. Fast stop DNA replication arrest at 44° |
PMID:16430690 PMID:8376347 |
Affects Tau but not Gamma | ||
|
dnaX36 |
Weak mutator in mismatch repair deficient strain |
PMID:16430690 |
|||||
|
dnaX0 |
|||||||
|
dnaX2016 |
G118D |
Growth Phenotype |
Ts for replication and growth |
PMID:8376347 |
|||
|
dnaX36(ts) |
temperature sensitive |
||||||
|
dnaXE145A |
E145A |
failure to suppress parE10 when overexpressed |
PMID:12535532 |
||||
|
dnaX(Ts) |
G118D |
Growth Phenotype |
temperature sensitive for replication and growth |
PMID:12775696 |
phenotype suppressed by mutations in DnaA | ||
| edit table |
<protect></protect>
Notes
The dnaX locus was identified by the J, Walker lab[5] as the site of one of several ts mutations in strain SG133, which was originally thought to contain a single mutation in dnaZ.[6]
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW0459 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAGTTATCAGGTCTTAGCCCG Primer 2:CCAATGGGGCGGATACTTTCTTC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 28% [8] | ||
|
Linked marker |
est. P1 cotransduction: 72% [8] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10245 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10245 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000239 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0241 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0001633 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
DnaX |
|---|---|
| Synonyms |
DNA polymerase III/DNA elongation factor III, tau and gamma subunits[1], DnaZ[2][1], B0470[2][1] , dnaZ, ECK0464, JW0459, b0470 |
| Product description |
|
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
DNA polymerase III
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0043846 |
DNA polymerase III, DnaX complex |
C |
Missing: evidence, reference | |||||
|
GO:0003689 |
DNA clamp loader activity |
F |
Missing: evidence, reference | |||||
|
GO:0000166 |
nucleotide binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003593 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000166 |
nucleotide binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003593 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008921 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008921 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003887 |
DNA-directed DNA polymerase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012763 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003887 |
DNA-directed DNA polymerase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012763 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003887 |
DNA-directed DNA polymerase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:2.7.7.7 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003887 |
DNA-directed DNA polymerase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:2.7.7.7 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003887 |
DNA-directed DNA polymerase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0239 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003887 |
DNA-directed DNA polymerase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0239 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005515 |
protein binding |
PMID:19703395 |
IPI: Inferred from Physical Interaction |
UniProtKB:P0A988 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005515 |
protein binding |
PMID:19703395 |
IPI: Inferred from Physical Interaction |
UniProtKB:P0A988 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001270 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001270 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003959 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003959 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012763 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012763 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006260 |
DNA replication |
PMID:391641 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0006260 |
DNA replication |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008921 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0017111 |
nucleoside-triphosphatase activity |
PMID:11029431 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0043846 |
DNA polymerase III, DnaX complex |
PMID:8376347 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0006260 |
DNA replication |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012763 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016887 |
ATPase activity |
PMID:2681183 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0006260 |
DNA replication |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0235 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030337 |
DNA polymerase processivity factor activity |
PMID:11525729 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0009360 |
DNA polymerase III complex |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012763 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030337 |
DNA polymerase processivity factor activity |
PMID:7494000 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0009360 |
DNA polymerase III complex |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR012763 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016740 |
transferase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0808 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016740 |
transferase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0808 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016779 |
nucleotidyltransferase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0548 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016779 |
nucleotidyltransferase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0548 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0017111 |
nucleoside-triphosphatase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003593 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0017111 |
nucleoside-triphosphatase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003593 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions
DNA polymerase III
See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of DNA polymerase III, preinitiation complex |
could be indirect |
||
|
Protein |
rplA |
PMID:15690043 |
Experiment(s):EBI-883400 | |
|
Protein |
fecB |
PMID:15690043 |
Experiment(s):EBI-890677 | |
|
Protein |
nusG |
PMID:15690043 |
Experiment(s):EBI-890677 | |
|
Protein |
rplL |
PMID:15690043 |
Experiment(s):EBI-890677 | |
|
Protein |
rpmC |
PMID:15690043 |
Experiment(s):EBI-890677 | |
|
Protein |
rpsF |
PMID:15690043 |
Experiment(s):EBI-890677 | |
|
Protein |
ugpB |
PMID:15690043 |
Experiment(s):EBI-890677 | |
|
Protein |
yfiF |
PMID:15690043 |
Experiment(s):EBI-890677 | |
|
Protein |
yjgD |
PMID:15690043 |
Experiment(s):EBI-890677 | |
|
Protein |
rhsD |
PMID:16606699 |
Experiment(s):EBI-1136784 | |
|
Protein |
rpoC |
PMID:16606699 |
Experiment(s):EBI-1136784 | |
|
Protein |
ugpB |
PMID:19402753 |
LCMS(ID Probability):99.0 | |
|
Protein |
fecB |
PMID:19402753 |
LCMS(ID Probability):99.0 | |
|
Protein |
Subunits of DNA polymerase III, holoenzyme |
could be indirect |
||
|
Protein |
ParC from Topo IV |
|
PMID:12535532 |
Microscopy- colocalization at the fork lost in dnaX mutant
|
|
Protein |
subunits of the clamp loader |
PMID:11525729 |
||
|
Protein |
DnaB (Helicase)-Interacts with tau |
PMID:10748120 |
||
|
Protein |
alpha subunit of DNA Polymerase III- interacts with tau |
PMID:10748120 |
| |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Quantitation of DnaX products in replisomes shows three molecules of Tau and no Gamma at the replication fork [9].
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MSYQVLARKW RPQTFADVVG QEHVLTALAN GLSLGRIHHA YLFSGTRGVG KTSIARLLAK GLNCETGITA TPCGVCDNCR EIEQGRFVDL IEIDAASRTK VEDTRDLLDN VQYAPARGRF KVYLIDEVHM LSRHSFNALL KTLEEPPEHV KFLLATTDPQ KLPVTILSRC LQFHLKALDV EQIRHQLEHI LNEEHIAHEP RALQLLARAA EGSLRDALSL TDQAIASGDG QVSTQAVSAM LGTLDDDQAL SLVEAMVEAN GERVMALINE AAARGIEWEA LLVEMLGLLH RIAMVQLSPA ALGNDMAAIE LRMRELARTI PPTDIQLYYQ TLLIGRKELP YAPDRRMGVE MTLLRALAFH PRMPLPEPEV PRQSFAPVAP TAVMTPTQVP PQPQSAPQQA PTVPLPETTS QVLAARQQLQ RVQGATKAKK SEPAAATRAR PVNNAALERL ASVTDRVQAR PVPSALEKAP AKKEAYRWKA TTPVMQQKEV VATPKALKKA LEHEKTPELA AKLAAEAIER DPWAAQVSQL SLPKLVEQVA LNAWKEESDN AVCLHLRSSQ RHLNNRGAQQ KLAEALSMLK GSTVELTIVE DDNPAVRTPL EWRQAIYEEK LAQARESIIA DNNIQTLRRF FDAELDEESI RPI |
| Length |
643 |
| Mol. Wt |
71.138 kDa |
| pI |
6.8 (calculated) |
| Extinction coefficient |
46,410 - 47,160 (calc based on 9 Y, 6 W, and 6 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0001633 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10245 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10245 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000239 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0241 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
dnaX encodes two gene products from the same gene. A programmed frameshift is needed to generate the gamma subunit[10][11][12].
The amount of gamma is estimated to be about 20 molecules per cell, based on an estimate of 10 dimers[13]. However, while gamma purifies as a dimer, it is present as one subunit in the τ2γδδ'χψ DnaX complex.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
Ecoli K-12 |
14.346+/-0.068 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.056447437 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
327 |
molecules/cell/generation |
|
Ribosome Profiling |
gamma isoform |
PMID: 24766808 |
|
Protein |
E. coli K-12 MG1655 |
84 |
molecules/cell/generation |
|
Ribosome Profiling |
gamma isoform |
PMID: 24766808 |
|
Protein |
E. coli K-12 MG1655 |
135 |
molecules/cell/generation |
|
Ribosome Profiling |
gamma isoform |
PMID: 24766808 |
|
Protein |
E. coli K-12 MG1655 |
196 |
molecules/cell/generation |
|
Ribosome Profiling |
tau isoform |
PMID: 24766808 |
|
Protein |
E. coli K-12 MG1655 |
85 |
molecules/cell/generation |
|
Ribosome Profiling |
tau isoform |
PMID: 24766808 |
|
Protein |
E. coli K-12 MG1655 |
134 |
molecules/cell/generation |
|
Ribosome Profiling |
tau isoform |
PMID: 24766808 |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
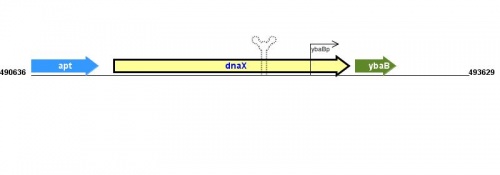 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:491296..491336
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for dnaX | |
|
microarray |
Summary of data for dnaX from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (491102..491347) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to dnaX Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10245 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0241 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10245 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000239 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0001633 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Shigella flexneri |
DNAX |
From SHIGELLACYC |
|
E. coli O157 |
DNAX |
From ECOO157CYC |
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Danio rerio |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Drosophila melanogaster |
|
From Inparanoid:20070104 |
|
Drosophila pseudoobscura |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Macaca mulatta |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Saccharomyces cerevisiae |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Tetraodon nigroviridis |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
PF00004 ATPase family associated with various cellular activities (AAA) |
||
|
PF12168 DNA polymerase III subunits tau domain IV DnaB-binding |
||
|
PF12169 DNA polymerase III subunits gamma and tau domain III |
||
|
PF12170 DNA polymerase III tau subunit V interacting with alpha |
||
|
EcoCyc:EG10245 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10245 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000239 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0241 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0001633 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 3.9 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ Henson, JM et al. (1979) Isolation and characterization of dnaX and dnaY temperature-sensitive mutants of Escherichia coli. Genetics 92 1041-59 PubMed
- ↑ Sevastopoulos, CG et al. (1977) Large-scale automated isolation of Escherichia coli mutants with thermosensitive DNA replication. Proc. Natl. Acad. Sci. U.S.A. 74 3485-9 PubMed
- ↑ 7.0 7.1 CGSC: The Coli Genetics Stock Center
- ↑ 8.0 8.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Reyes-Lamothe, R et al. (2010) Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 328 498-501 PubMed
- ↑ Tsuchihashi, Z & Kornberg, A (1990) Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 87 2516-20 PubMed
- ↑ Blinkowa, AL & Walker, JR (1990) Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res. 18 1725-9 PubMed
- ↑ Flower, AM & McHenry, CS (1990) The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl. Acad. Sci. U.S.A. 87 3713-7 PubMed
- ↑ Hübscher, U & Kornberg, A (1980) The dnaZ protein, the gamma subunit of DNA polymerase III holoenzyme of Escherichia coli. J. Biol. Chem. 255 11698-703 PubMed
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories


