dnaX:Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Overview | Transcription | Translation | Turnover | Experimental | Links | References | Suggestions |
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
dnaX encodes two gene products from the same gene. A programmed frameshift is needed to generate the gamma subunit[1][2][3].
The amount of gamma is estimated to be about 20 molecules per cell, based on an estimate of 10 dimers[4]. However, while gamma purifies as a dimer, it is present as one subunit in the τ2γδδ'χψ DnaX complex.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
Ecoli K-12 |
14.346+/-0.068 |
Molecules/cell |
|
Single Molecule Fluorescence |
||
|
mRNA |
Ecoli K-12 |
0.056447437 |
Molecules/cell |
|
by RNA_Seq |
||
|
Protein |
E. coli K-12 MG1655 |
327 |
molecules/cell/generation |
|
Ribosome Profiling |
gamma isoform |
|
|
Protein |
E. coli K-12 MG1655 |
84 |
molecules/cell/generation |
|
Ribosome Profiling |
gamma isoform |
|
|
Protein |
E. coli K-12 MG1655 |
135 |
molecules/cell/generation |
|
Ribosome Profiling |
gamma isoform |
|
|
Protein |
E. coli K-12 MG1655 |
196 |
molecules/cell/generation |
|
Ribosome Profiling |
tau isoform |
|
|
Protein |
E. coli K-12 MG1655 |
85 |
molecules/cell/generation |
|
Ribosome Profiling |
tau isoform |
|
|
Protein |
E. coli K-12 MG1655 |
134 |
molecules/cell/generation |
|
Ribosome Profiling |
tau isoform |
|
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
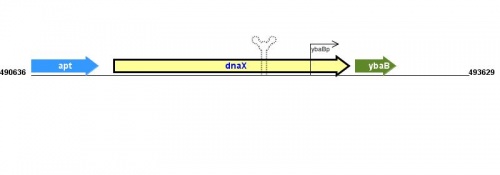 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:491296..491336
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for dnaX | |
|
microarray |
Summary of data for dnaX from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (491102..491347) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to dnaX Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ Tsuchihashi, Z & Kornberg, A (1990) Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 87 2516-20 PubMed
- ↑ Blinkowa, AL & Walker, JR (1990) Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res. 18 1725-9 PubMed
- ↑ Flower, AM & McHenry, CS (1990) The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl. Acad. Sci. U.S.A. 87 3713-7 PubMed
- ↑ Hübscher, U & Kornberg, A (1980) The dnaZ protein, the gamma subunit of DNA polymerase III holoenzyme of Escherichia coli. J. Biol. Chem. 255 11698-703 PubMed
- ↑ 5.0 5.1 Taniguchi, Y et al. (2010) Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329 533-8 PubMed
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 Li, GW et al. (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157 624-35 PubMed
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories