cyaA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
cyaA |
|---|---|
| Gene Synonym(s) |
ECK3800, b3806, JW3778, cya[1], cya |
| Product Desc. |
Adenylate cyclase[2] |
| Product Synonyms(s) |
adenylate cyclase[1], B3806[3][1], Cya[3][1], CyaA[3][1] , cya, ECK3800, JW3778, b3806 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Polyamine modulon.[2]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
cyaA |
|---|---|
| Mnemonic |
Cyclase, adenylate |
| Synonyms |
ECK3800, b3806, JW3778, cya[1], cya |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
85.98 minutes |
MG1655: 3989176..3991722 |
||
|
NC_012967: 3952983..3955529 |
||||
|
NC_012759: 3878845..3881391 |
||||
|
W3110 |
|
W3110: 3645528..3642982 |
||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
cyaA (Keio:JW3778) |
deletion |
deletion |
PMID:16738554 |
||||
|
cyaA::Tn5KAN-I-SceI (FB21464) |
Insertion at nt 1522 in Minus orientation |
PMID:15262929 |
does not contain pKD46 | ||||
|
cyaA::Tn5KAN-I-SceI (FB21465) |
Insertion at nt 1522 in Minus orientation |
PMID:15262929 |
contains pKD46 | ||||
|
cyaA2(del) |
PMID:4583241 PMID:2022622 |
||||||
|
cyaA854(del) |
PMID:2670905 |
||||||
|
cyaA1 |
PMID:173710 |
||||||
|
cyaA1400(del)::kan |
PMID:2022622 |
||||||
|
cyaA201(del) |
PMID:4583241 PMID:2022622 |
||||||
|
cyaA1404(del)::FRT |
PMID:10829079 |
||||||
|
cyaA1403(del)::kan |
PMID:10829079 |
||||||
|
cyaA161 |
PMID:9495769 |
||||||
|
cyaA751(del)::kan |
PMID:16738554 |
||||||
|
cya(del) |
deletion |
deletion |
Auxotrophies |
Mutation inhibits stationary phase SOS induction of the yebG gene (i.e. dependent on intracellular cAMP levels), fig 3. |
PMID:10474193 |
||
|
cya in strain L176 |
Sensitivity to |
sensitivity to streptozotocin |
PMID:6450313 |
strain L176 |
figure 3 | ||
|
cya (del) crp (del) |
Sensitivity to |
sensitivity to streptozotocin |
PMID:6450313 |
Strain: CA8445 |
table 2 and figure 3 | ||
|
cya crp* relA + |
Resistant to |
crp allele that modified cyclic AMP receptor proteins, which acts independently of cyclic AMP, is resistant to smg (serine+methionine+glycine) in a relA background. |
PMID:230407 |
Experimental strain: BM6163 |
table 4 | ||
|
cya(del) |
Resistant to |
Resistance to UV irridiation (254nm). |
PMID:1379686 |
Strain: GE1068 |
See figure 1 | ||
|
Salmonella typhimurium LT2 cya-deficient |
Resistant to |
partially resistant to 22 antibiotics (including fosfomycin, nalidixic acid, and streptomycin) |
PMID:201606 |
||||
|
cya in strain NCR-C57 |
Resistant to |
Resistance to T6 Phage |
PMID:227838 |
Experiential Strain: NCR-C57 Parental Strain: NCR-701 |
Table 3 | ||
|
cyaA751(del)::FRTKanFRT |
deletion |
Mutagenesis Rate |
Decrease in Stress Induced Mutagenesis (SIM). |
PMID:23224554 |
Parent Strain: 23224554 Experimental Strain: SMR11998 |
The mutation conferred a significant decrease in SIM with a reduction in mutant frequency by 67 to 89 percent. See table S3 for full experimental data. | |
|
cya-408 |
Resistant to |
Resistant to Fosfomycin |
PMID:201606 |
Strain: TA3301 |
Strain Parent: LT2 Strain donor: TA2301 | ||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW3778 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCTACCTCTATATTGAGACTCT Primer 2:CCaGAAAAATATTGCTGTAATAG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 23% [5] | ||
|
metE3079::Tn10 |
Linked marker |
est. P1 cotransduction: 49% [5] | |
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10170 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10170 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000166 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0168 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012432 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
CyaA |
|---|---|
| Synonyms |
adenylate cyclase[1], B3806[3][1], Cya[3][1], CyaA[3][1] , cya, ECK3800, JW3778, b3806 |
| Product description |
Adenylate cyclase[2] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004016 |
adenylate cyclase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000274 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004016 |
adenylate cyclase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:4.6.1.1 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0963 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0086 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006171 |
cAMP biosynthetic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000274 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006171 |
cAMP biosynthetic process |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0115 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
dnaE |
PMID:16606699 |
Experiment(s):EBI-1146708 | |
|
Protein |
rapA |
PMID:16606699 |
Experiment(s):EBI-1146708 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MYLYIETLKQ RLDAINQLRV DRALAAMGPA FQQVYSLLPT LLHYHHPLMP GYLDGNVPKG ICLYTPDETQ RHYLNELELY RGMSVQDPPK GELPITGVYT MGSTSSVGQS CSSDLDIWVC HQSWLDSEER QLLQRKCSLL ENWAASLGVE VSFFLIDENR FRHNESGSLG GEDCGSTQHI LLLDEFYRTA VRLAGKRILW NMVPCDEEEH YDDYVMTLYA QGVLTPNEWL DLGGLSSLSA EEYFGASLWQ LYKSIDSPYK AVLKTLLLEA YSWEYPNPRL LAKDIKQRLH DGEIVSFGLD PYCMMLERVT EYLTAIEDFT RLDLVRRCFY LKVCEKLSRE RACVGWRRAV LSQLVSEWGW DEARLAMLDN RANWKIDQVR EAHNELLDAM MQSYRNLIRF ARRNNLSVSA SPQDIGVLTR KLYAAFEALP GKVTLVNPQI SPDLSEPNLT FIYVPPGRAN RSGWYLYNRA PNIESIISHQ PLEYNRYLNK LVAWAWFNGL LTSRTRLYIK GNGIVDLPKL QEMVADVSHH FPLRLPAPTP KALYSPCEIR HLAIIVNLEY DPTAAFRNQV VHFDFRKLDV FSFGENQNCL VGSVDLLYRN SWNEVRTLHF NGEQSMIEAL KTILGKMHQD AAPPDSVEVF CYSQHLRGLI RTRVQQLVSE CIELRLSSTR QETGRFKALR VSGQTWGLFF ERLNVSVQKL ENAIEFYGAI SHNKLHGLSV QVETNHVKLP AVVDGFASEG IIQFFFEETQ DENGFNIYIL DESNRVEVYH HCEGSKEELV RDVSRFYSSS HDRFTYGSSF INFNLPQFYQ IVKVDGREQV IPFRTKSIGN MPPANQDHDT PLLQQYFS |
| Length |
848 |
| Mol. Wt |
97.587 kDa |
| pI |
6.2 (calculated) |
| Extinction coefficient |
147,600 - 149,475 (calc based on 40 Y, 16 W, and 15 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0012432 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10170 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10170 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000166 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0168 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
372 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
263 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
342 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
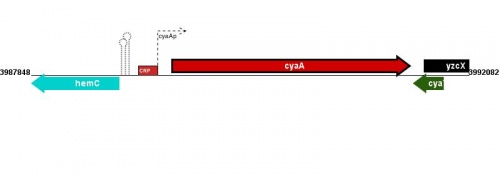 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:3989156..3989196
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for cyaA | |
|
microarray |
Summary of data for cyaA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (3988326..3988849) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to cyaA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10170 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0168 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10170 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000166 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012432 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Shigella flexneri |
CYAA |
From SHIGELLACYC |
|
E. coli O157 |
CYAA |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10170 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10170 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000166 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0168 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012432 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 CGSC: The Coli Genetics Stock Center
- ↑ 5.0 5.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories


