bamA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
bamA |
|---|---|
| Gene Synonym(s) |
ECK0176, b0177, JW0172, up05, ecfK, yzzN, yzzY, omp85(N.m.), bamA, yaeT[1][2][3], omp85, yaeT |
| Product Desc. |
YaeT[2]; YaeT-component of outer membrane protein assembly complex[4]; Component of Outer Membrane Protein Assembly Complex[4] |
| Product Synonyms(s) |
conserved protein[1], B0177[2][1], YzzN[2][1], YzzY[2][1], EcfK[2][1], YaeT[2][1], BamA [3], ecfK, ECK0176, JW0172, omp85, yaeT, yzzN, yzzY, b0177 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
A consensus has been reached for renaming the genes that encode components of the outer membrane β-barrel assembly machine, (formerly known as the YaeT complex of E. coli or the Omp85 complex of N. meningitidis). The new designation for this multi-component structure is the Bam complex (for beta-barrel assembly machine).
The Bam complex in E. coli consists of BamA (YaeT), BamB (YfgL), BamC (NlpB), BamD (YfiO), and BamE (SmpA). Letters have been assigned on the basis of molecular weight, from highest to lowest [3].
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
bamA |
|---|---|
| Mnemonic |
Beta-barrel assembly machine |
| Synonyms |
ECK0176, b0177, JW0172, up05, ecfK, yzzN, yzzY, omp85(N.m.), bamA, yaeT[1][2][3], omp85, yaeT |
| edit table |
</protect>
Notes
A consensus has been reached for renaming the genes that encode components of the outer membrane β-barrel assembly machine, (formerly known as the YaeT complex of E. coli or the Omp85 complex of N. meningitidis). The new designation for this multi-component structure is the Bam complex (for beta-barrel assembly machine).
The Bam complex in E. coli consists of BamA (YaeT), BamB (YfgL), BamC (NlpB), BamD (YfiO), and BamE (SmpA). Letters have been assigned on the basis of molecular weight, from highest to lowest [3].
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
4.27 minutes |
MG1655: 197928..200360 |
||
|
NC_012967: 200770..203202 |
||||
|
NC_012759: 197927..200359 |
||||
|
W3110 |
|
W3110: 197928..200360 |
||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
197988 |
Edman degradation |
PMID:9298646 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
yaeT6 |
in-frame insertion of Gln–Lys after amino acid 218 of YaeT (BamA) |
Sensitivity to |
Rifampicin, Erythromycin, Novobiocin, Bacitracin |
Suppressor of imp4213 sensitivity to deoxycholate and chenodeoxycholate | |||
| edit table |
<protect></protect>
Notes
yaeT6 carries a 6-bp in-frame insertion (agaaac) between codons 218 and 219 of yaeT. At the protein level, the suppressor mutation results in an in-frame insertion of Gln–Lys after amino acid 218 of YaeT. The yaeT6 mutation was isolated as a suppressor of the imp4213 allele of lptD.[5]
The mutant allele bamA66 is a deletion of the R64 codon of bamA. The mutant protein is referred to as BamA(delR64). The bamA66 mutation was isolated as a suppressor that conferred increased resistance to bacitracin of a strain carrying the imp208 allele of lptD. In an lptD+ background, bamA66 displays a slight sensitivity to vancomycin and rifampin and causes a moderate reduction (20–30%) in OMP levels, but is synthetically lethal with the depletion of BamB, conditionally lethal in the absence of DegP or SurA; i.e. no growth at or above 37°C and 40°C respectively.[6]
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW0172 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCGCGATGAAAAAGTTGCTCAT Primer 2:CCCCAGGTTTTACCGATGTTAAA | |
|
Linked marker |
est. P1 cotransduction: 10% [8] | ||
|
Linked marker |
est. P1 cotransduction: 25% [8] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G6093 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12676 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120002713 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2541 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000605 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
BamA |
|---|---|
| Synonyms |
conserved protein[1], B0177[2][1], YzzN[2][1], YzzY[2][1], EcfK[2][1], YaeT[2][1], BamA [3], ecfK, ECK0176, JW0172, omp85, yaeT, yzzN, yzzY, b0177 |
| Product description |
YaeT[2]; YaeT-component of outer membrane protein assembly complex[4]; Component of Outer Membrane Protein Assembly Complex[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0043165 |
Gram-negative-bacterium-type cell outer membrane assembly |
PMID:15851030 |
IPI: Inferred from Physical Interaction |
EcoliWiki:yfgL EcoliWiki:nlpB EcoliWiki:yfiO
|
P |
complete | ||
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-1003 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009279 |
cell outer membrane |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_01430 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005515 |
protein binding |
PMID:15851030 |
IPI: Inferred from Physical Interaction |
EcoliWiki:nlpB EcoliWiki:yfgL EcoliWiki:yfiO
|
F |
complete | ||
|
GO:0009279 |
cell outer membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0998 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009279 |
cell outer membrane |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0040 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of Outer Membrane Protein Assembly Complex |
|||
|
Protein |
skp |
PMID:19402753 |
LCMS(ID Probability):99.5 | |
|
Protein |
edd |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
gldA |
PMID:19402753 |
LCMS(ID Probability):99.4 | |
|
Protein |
yegQ |
PMID:19402753 |
MALDI(Z-score):27.086609 | |
|
Protein |
fur |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
acnB |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
gcvP |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
gabD |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
deoA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
aldA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
folD |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
BamB (yfgL) |
BamB directly binds BamA (YaeT). Interaction between BamA-BamB is independent of BamA-BamCDE interactions. [9],[10] |
||
|
Protein |
BamC (nlpB) |
BamC is part of the BamCDE sub-complex that binds to BamA (YaeT). Interaction of BamCDE sub-complex with BamA is independent of BamA-BamB interaction. [9],[10] |
||
|
Protein |
BamD (yfiO) |
BamD is part of the BamCDE sub-complex that binds to BamA (YaeT). BamD directly binds BamA. Interaction of BamCDE sub-complex with BamA is independent of BamA-BamB interaction. [9],[10] |
||
|
Protein |
BamE (smpA) |
BamE is part of the BamCDE sub-complex that binds to BamA (YaeT). Interaction of BamCDE sub-complex with BamA is independent of BamA-BamB interaction. [10] |
| |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes | |
|---|---|---|---|---|---|
|
outer membrane |
|||||
|
Outer Membrane |
PMID:15951436, PMID:16102012, PMID:9298646 |
||||
|
Outer Membrane |
PMID:9298646 |
| |||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MAMKKLLIAS LLFSSATVYG AEGFVVKDIH FEGLQRVAVG AALLSMPVRT GDTVNDEDIS NTIRALFATG NFEDVRVLRD GDTLLVQVKE RPTIASITFS GNKSVKDDML KQNLEASGVR VGESLDRTTI ADIEKGLEDF YYSVGKYSAS VKAVVTPLPR NRVDLKLVFQ EGVSAEIQQI NIVGNHAFTT DELISHFQLR DEVPWWNVVG DRKYQKQKLA GDLETLRSYY LDRGYARFNI DSTQVSLTPD KKGIYVTVNI TEGDQYKLSG VEVSGNLAGH SAEIEQLTKI EPGELYNGTK VTKMEDDIKK LLGRYGYAYP RVQSMPEIND ADKTVKLRVN VDAGNRFYVR KIRFEGNDTS KDAVLRREMR QMEGAWLGSD LVDQGKERLN RLGFFETVDT DTQRVPGSPD QVDVVYKVKE RNTGSFNFGI GYGTESGVSF QAGVQQDNWL GTGYAVGING TKNDYQTYAE LSVTNPYFTV DGVSLGGRLF YNDFQADDAD LSDYTNKSYG TDVTLGFPIN EYNSLRAGLG YVHNSLSNMQ PQVAMWRYLY SMGEHPSTSD QDNSFKTDDF TFNYGWTYNK LDRGYFPTDG SRVNLTGKVT IPGSDNEYYK VTLDTATYVP IDDDHKWVVL GRTRWGYGDG LGGKEMPFYE NFYAGGSSTV RGFQSNTIGP KAVYFPHQAS NYDPDYDYEC ATQDGAKDLC KSDDAVGGNA MAVASLEFIT PTPFISDKYA NSVRTSFFWD MGTVWDTNWD SSQYSGYPDY SDPSNIRMSA GIALQWMSPL GPLVFSYAQP FKKYDGDKAE QFQFNIGKTW |
| Length |
810 |
| Mol. Wt |
90.553 kDa |
| pI |
4.8 (calculated) |
| Extinction coefficient |
141,530 - 141,780 (calc based on 47 Y, 13 W, and 2 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
</protect>
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0000605 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:G6093 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12676 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120002713 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2541 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 EMG2 |
180 |
molecules/cell |
|
Quantitative protein sequencing |
Spot ID: M60 |
PMID: 9298646 |
|
Protein |
E. coli K-12 EMG2 |
80 |
molecules/cell |
|
Quantitative protein sequencing |
Spot ID: M61 |
PMID: 9298646 |
|
Protein |
E. coli K-12 MG1655 |
3904 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
1512 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
2826 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
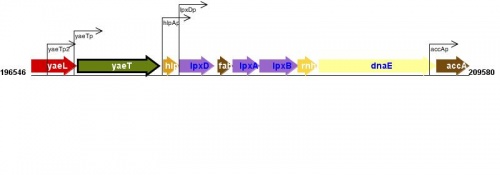 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:197908..197948
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for yaeT | |
|
microarray |
Summary of data for yaeT from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (197801..197953) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to bamA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G6093 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2541 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12676 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120002713 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000605 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Danio rerio |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Tetraodon nigroviridis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
YAET |
From SHIGELLACYC |
|
E. coli O157 |
YAET |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G6093 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12676 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120002713 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2541 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000605 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 4.0 4.1 4.2 4.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 5.0 5.1 Ruiz, N et al. (2006) Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem. Biol. 1 385-95 PubMed
- ↑ Bennion, D et al. (2010) Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol. Microbiol. 77 1153-71 PubMed
- ↑ 7.0 7.1 CGSC: The Coli Genetics Stock Center
- ↑ 8.0 8.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ 9.0 9.1 9.2 Malinverni, JC et al. (2006) YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61 151-64 PubMed
- ↑ 10.0 10.1 10.2 10.3 Sklar, JG et al. (2007) Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 104 6400-5 PubMed
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories
- Genes in OpenBioSystems with Promoter Fusions
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Bos taurus
- Genes with homologs in Canis familiaris
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Danio rerio
- Genes with homologs in Gallus gallus
- Genes with homologs in Homo sapiens
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Schizosaccharomyces pombe
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Tetraodon nigroviridis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


