ytfN:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
ytfN |
|---|---|
| Gene Synonym(s) |
ECK4217, b4221, JW4180, ytfO[1], ytfO |
| Product Desc. |
Function unknown[4] |
| Product Synonyms(s) |
conserved protein[1], B4221[2][1], YtfO[2][1], YtfN[2][1] , ECK4217, JW4180, ytfO, b4221 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
May have signal peptide (weak prediction), or an uncleaved signal anchor (predicted single N-terminal TM region). Putative ytfMNP operon.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
ytfN |
|---|---|
| Mnemonic |
Systematic nomenclature |
| Synonyms |
ECK4217, b4221, JW4180, ytfO[1], ytfO |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
95.74 minutes |
MG1655: 4442135..4445914 |
||
|
NC_012967: 4427869..4431648 |
||||
|
NC_012759: 4380870..4384649 |
||||
|
W3110 |
|
W3110: 4448792..4452571 |
||
|
DH10B: 4542497..4546276 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔytfN (Keio:JW4180) |
deletion |
deletion |
PMID:16738554 |
||||
|
zjf-920::Tn10 |
Insertion at 4,442,823 bp in MG1655 (NC_000913) |
adapted from Nichols et al.[6] |
Synonyms: zjh-901::Tn10, zjg-920::Tn10 | ||||
|
ΔytfN770::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW4180 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAGTTTATGGAAAAAAATCAG Primer 2:CCAAACTCGAACTGATAGAGCAA | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 57% [6] | ||
|
Linked marker |
est. P1 cotransduction: % [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB: |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE: |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP: |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
YtfN |
|---|---|
| Synonyms |
conserved protein[1], B4221[2][1], YtfO[2][1], YtfN[2][1] , ECK4217, JW4180, ytfO, b4221 |
| Product description |
Function unknown[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
yoaB |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MSLWKKISLG VVIVILLLLG SVAFLVGTTS GLHLVFKAAD RWVPGLDIGK VTGGWRDLTL SDVRYEQPGV AVKAGNLHLA VGLECLWNSS VCINDLALKD IQVNIDSKKM PPSEQVEEEE DSGPLDLSTP YPITLTRVAL DNVNIKIDDT TVSVMDFTSG LNWQEKTLTL KPTSLKGLLI ALPKVAEVAQ EEVVEPKIEN PQPDEKPLGE TLKDLFSRPV LPEMTDVHLP LNLNIEEFKG EQLRVTGDTD ITVSTMLLKV SSIDGNTKLD ALDIDSSQGI VNASGTAQLS DNWPVDITLN STLNVEPLKG EKVKLKMGGA LREQLEIGVN LSGPVDMDLR AQTRLAEAGL PLNVEVNSKQ LYWPFTGEKQ YQADDLKLKL TGKMTDYTLS MRTAVKGQEI PPATITLDAK GNEQQVNLDK LTVAALEGKT ELKALLDWQQ AISWRGELTL NGINTAKEFP DWPSKLNGLI KTRGSLYGGT WQMDVPELKL TGNVKQNKVN VDGTLKGNSY MQWMIPGLHL ELGPNSAEVK GELGVKDLNL DATINAPGLD NALPGLGGTA KGLVKVRGTV EAPQLLADIT ARGLRWQELS VAQVRVEGDI KSTDQIAGKL DVRVEQISQP DVNINLVTLN AKGSEKQHEL QLRIQGEPVS GQLNLAGSFD RKEERWKGTL SNTRFQTPVG PWSLTRDIAL DYRNKEQKIS IGPHCWLNPN AELCVPQTID AGAEGRAVVN LNRFDLAMLK PFMPETTQAS GIFTGKADVA WDTTKEGLPQ GSITLSGRNV QVTQTVNDAA LPVAFQTLNL TAELRNNRAE LGWTIRLTNN GQFDGQVQVT DPQGRRNLGG NVNIRNFNLA MINPIFTRGE KAAGMVSANL RLGGDVQSPQ LFGQLQVTGV DIDGNFMPFD MQPSQLAVNF NGMRSTLAGT VRTQQGEIYL NGDADWSQIE NWRARVTAKG SKVRITVPPM VRMDVSPDVV FEATPNLFTL DGRVDVPWAR IVVHDLPESA VGVSSDVVML NDNLQPEEPK TASIPINSNL IVHVGNNVRI DAFGLKARLT GDLNVVQDKQ GLGLNGQINI PEGRFHAYGQ DLIVRKGELL FSGPPDQPYL NIEAIRNPDA TEDDVIAGVR VTGLADEPKA EIFSDPAMSQ QAALSYLLRG QGLESDQSDS AAMTSMLIGL GVAQSGQIVG KIGETFGVSN LALDTQGVGD SSQVVVSGYV LPGLQVKYGV GIFDSIATLT LRYRLMPKLY LEAVSGVDQA LDLLYQFEF |
| Length |
1,259 |
| Mol. Wt |
136.781 kDa |
| pI |
4.8 (calculated) |
| Extinction coefficient |
140,830 - 141,330 (calc based on 17 Y, 21 W, and 4 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB: |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE: |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
1.04E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
0.617+/-0.048 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.032283673 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
127 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
54 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
68 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
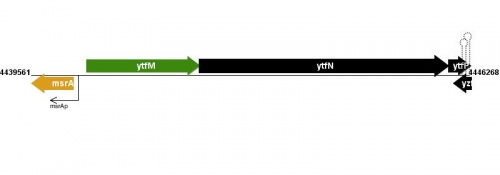 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:4442115..4442155
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for ytfN | |
|
microarray |
Summary of data for ytfN from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to ytfN Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
RegulonDB: |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP: |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
YTFN |
From SHIGELLACYC |
|
E. coli O157 |
YTFN |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc: |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene: |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB: |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE: |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP: |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 5.3 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 6.2 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories
