ydiJ:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
ydiJ |
|---|---|
| Gene Synonym(s) |
ECK1684, b1687, JW1677[1], JW1677 |
| Product Desc. |
predicted FAD-linked oxidoreductase[2][3] Function unknown[4] |
| Product Synonyms(s) |
predicted FAD-linked oxidoreductase[1], B1687[2][1], YdiJ[2][1] , ECK1684, JW1677, b1687 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Overexpression causes abnormal biofilm architecture, increased crystal violet staining, and reduced motility. YdiJ is homologous to glycolate oxidase. P. putida ydiJ mutant is unaffected for growth and survival.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
ydiJ |
|---|---|
| Mnemonic |
Systematic nomenclature |
| Synonyms |
ECK1684, b1687, JW1677[1], JW1677 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
38.01 minutes |
MG1655: 1766709..1763653 |
||
|
NC_012967: 1745952..1742896 |
||||
|
NC_012759: 1655712..1658768 |
||||
|
W3110 |
|
W3110: 1770399..1767343 |
||
|
DH10B: 1857280..1854224 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔydiJ (Keio:JW1677) |
deletion |
deletion |
PMID:16738554 |
||||
|
ΔydiJ::kan |
deletion |
Biolog:respiration |
unable to respire L-Asparagine |
PMID:16095938 |
|||
|
ΔydiJ::kan |
deletion |
Biolog:respiration |
unable to respire L-Aspartate |
PMID:16095938 |
|||
|
ΔydiJ762::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW1677 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCATTCCACAGATTTCCCAGGC Primer 2:CCTTTAATAATCTCCAGTAAAGC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: % [6] | ||
|
zdi-925::Tn10 |
Linked marker |
est. P1 cotransduction: 63% [6] | |
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G6913 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG13969 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120003501 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB3726 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0005632 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
YdiJ |
|---|---|
| Synonyms |
predicted FAD-linked oxidoreductase[1], B1687[2][1], YdiJ[2][1] , ECK1684, JW1677, b1687 |
| Product description |
predicted FAD-linked oxidoreductase[2][3] Function unknown[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0003824 |
catalytic activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004113 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003824 |
catalytic activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR016164 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003824 |
catalytic activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR016166 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003824 |
catalytic activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR016167 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009055 |
electron carrier activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR017900 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016491 |
oxidoreductase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006094 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0050660 |
FAD binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004113 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0050660 |
FAD binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006094 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0050660 |
FAD binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR016164 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0050660 |
FAD binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR016166 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0050660 |
FAD binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR016167 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0051536 |
iron-sulfur cluster binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR017900 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
ydfG |
PMID:19402753 |
MALDI(Z-score):21.453935 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MIPQISQAPG VVQLVLNFLQ ELEQQGFTGD TATSYADRLT MSTDNSIYQL LPDAVVFPRS TADVALIARL AAQERYSSLI FTPRGGGTGT NGQALNQGII VDMSRHMNRI IEINPEEGWV RVEAGVIKDQ LNQYLKPFGY FFAPELSTSN RATLGGMINT DASGQGSLVY GKTSDHVLGV RAVLLGGDIL DTQPLPVELA ETLGKSNTTI GRIYNTVYQR CRQQRQLIID NFPKLNRFLT GYDLRHVFND EMTEFDLTRI LTGSEGTLAF ITEARLDITR LPKVRRLVNV KYDSFDSALR NAPFMVEARA LSVETVDSKV LNLAREDIVW HSVSELITDV PDQEMLGLNI VEFAGDDEAL IDERVNALCA RLDELIASHQ AGVIGWQVCR ELAGVERIYA MRKKAVGLLG NAKGAAKPIP FAEDTCVPPE HLADYIAEFR ALLDSHGLSY GMFGHVDAGV LHVRPALDMC DPQQEILMKQ ISDDVVALTA KYGGLLWGEH GKGFRAEYSP AFFGEELFAE LRKVKAAFDP HNRLNPGKIC PPEGLDAPMM KVDAVKRGTF DRQIPIAVRQ QWRGAMECNG NGLCFNFDAR SPMCPSMKIT QNRIHSPKGR ATLVREWLRL LADRGVDPLK LEQELPESGV SLRTLIARTR NSWHANKGEY DFSHEVKEAM SGCLACKACS TQCPIKIDVP EFRSRFLQLY HTRYLRPLRD HLVATVESYA PLMARAPKTF NFFINQPLVR KLSEKHIGMV DLPLLSVPSL QQQMVGHRSA NMTLEQLESL NAEQKARTVL VVQDPFTSYY DAQVVADFVR LVEKLGFQPV LLPFSPNGKA QHIKGFLNRF AKTAKKTADF LNRMAKLGMP MVGVDPALVL CYRDEYKLAL GEERGEFNVL LANEWLASAL ESQPVATVSG ESWYFFGHCT EVTALPGAPA QWAAIFARFG AKLENVSVGC CGMAGTYGHE AKNHENSLGI YELSWHQAMQ RLPRNRCLAT GYSCRSQVKR VEGTGVRHPV QALLEIIK |
| Length |
1,018 |
| Mol. Wt |
113.249 kDa |
| pI |
7.1 (calculated) |
| Extinction coefficient |
100,730 - 103,105 (calc based on 27 Y, 11 W, and 19 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0005632 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:G6913 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG13969 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120003501 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB3726 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
Ecoli K-12 |
6.558+/-0.137 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.073134817 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
728 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
897 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
620 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
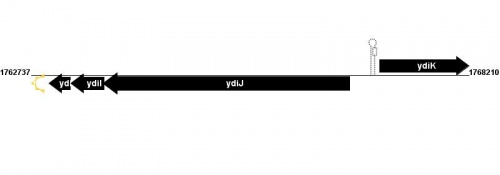 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:1766689..1766729
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for ydiJ | |
|
microarray |
Summary of data for ydiJ from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to ydiJ Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G6913 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB3726 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG13969 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120003501 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0005632 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Shigella flexneri |
YDIJ |
From SHIGELLACYC |
|
E. coli O157 |
YDIJ |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G6913 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG13969 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120003501 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB3726 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0005632 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories


