torA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
torA |
|---|---|
| Gene Synonym(s) |
ECK0988, b0997, JW0982[1], JW0982 |
| Product Desc. |
trimethylamine N-oxide reductase, catalytic subunit[2][3]; Component of trimethylamine N-oxide reductase I[2][3] Molybdoprotein trimethylamine N-oxide reductase[4] |
| Product Synonyms(s) |
trimethylamine N-oxide (TMAO) reductase I, catalytic subunit[1], B0997[2][1], TorA[2][1] , ECK0988, JW0982, b0997 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Verified Tat substrate: TorA has a Tat/Sec (Class II) 39 aa signal peptide (Tullman-Ercek, 2007).[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
torA |
|---|---|
| Mnemonic |
Trimethylamine oxide reductase |
| Synonyms |
ECK0988, b0997, JW0982[1], JW0982 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
22.81 minutes |
MG1655: 1058479..1061025 |
||
|
NC_012967: 1075546..1078092 |
||||
|
NC_012759: 961447..963993 |
||||
|
W3110 |
|
W3110: 1059678..1062224 |
||
|
DH10B: 1112407..1114953 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
1058596 |
Edman degradation |
PMID:8039676 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔtorA (Keio:JW0982) |
deletion |
deletion |
PMID:16738554 |
||||
|
torA::Tn5KAN-2 (FB20082) |
Insertion at nt 973 in Minus orientation |
PMID:15262929 |
does not contain pKD46 | ||||
|
ΔtorA741::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW0982 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAACAATAACGATCTCTTTCA Primer 2:CCTGATTTCACCTGCGACGCGGG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 6% [6] | ||
|
Linked marker |
est. P1 cotransduction: 88% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11814 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11814 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001747 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB1761 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0003372 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
TorA |
|---|---|
| Synonyms |
trimethylamine N-oxide (TMAO) reductase I, catalytic subunit[1], B0997[2][1], TorA[2][1] , ECK0988, JW0982, b0997 |
| Product description |
trimethylamine N-oxide reductase, catalytic subunit[2][3]; Component of trimethylamine N-oxide reductase I[2][3] Molybdoprotein trimethylamine N-oxide reductase[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0030288 |
outer membrane-bounded periplasmic space |
C |
Seeded from Riley et al 2006 [1]. |
Missing: evidence, reference | ||||
|
GO:0009055 |
electron carrier activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006655 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009055 |
electron carrier activity |
PMID:11056172 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0009060 |
aerobic respiration |
PMID:17850256 |
IEP: Inferred from Expression Pattern |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0009061 |
anaerobic respiration |
PMID:17850256 |
IEP: Inferred from Expression Pattern |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0016491 |
oxidoreductase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006655 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016491 |
oxidoreductase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006656 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016491 |
oxidoreductase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006657 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016491 |
oxidoreductase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006658 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016491 |
oxidoreductase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0560 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030151 |
molybdenum ion binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006657 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030151 |
molybdenum ion binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011887 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030151 |
molybdenum ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0500 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030151 |
molybdenum ion binding |
PMID:18163615 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0042597 |
periplasmic space |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011887 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0042597 |
periplasmic space |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0574 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0042597 |
periplasmic space |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0200 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0042597 |
periplasmic space |
PMID:2512991 |
IDA: Inferred from Direct Assay |
C |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0050626 |
trimethylamine-N-oxide reductase (cytochrome c) activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011887 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0050626 |
trimethylamine-N-oxide reductase (cytochrome c) activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:1.7.2.3 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006658 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011887 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0560 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0050626 |
trimethylamine-N-oxide reductase (cytochrome c) activity |
PMID:18522945 |
IMP: Inferred from Mutant Phenotype |
F |
complete | |||
|
GO:0005515 |
protein binding |
PMID:11562502 |
IPI: Inferred from Physical Interaction |
EcoliWiki:torC
|
F |
complete | ||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of trimethylamine N-oxide reductase I |
could be indirect |
||
|
Protein |
yibN |
PMID:16606699 |
Experiment(s):EBI-1138562 | |
|
Protein |
mutT |
PMID:16606699 |
Experiment(s):EBI-1138562 | |
|
Protein |
hns |
PMID:16606699 |
Experiment(s):EBI-1138562 | |
|
Protein |
yghW |
PMID:16606699 |
Experiment(s):EBI-1138562 | |
|
Protein |
napA |
PMID:16606699 |
Experiment(s):EBI-1138562 | |
|
Protein |
tatE |
PMID:16606699 |
Experiment(s):EBI-1138562 | |
|
Protein |
groL |
PMID:16606699 |
Experiment(s):EBI-1138562 | |
|
Protein |
ybfO |
PMID:16606699 |
Experiment(s):EBI-1138562 | |
|
Protein |
metL |
PMID:16606699 |
Experiment(s):EBI-1138562 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
periplasm |
||||
|
Periplasm |
PMID:15073303 |
| ||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MNNNDLFQAS RRRFLAQLGG LTVAGMLGPS LLTPRRATAA QAATDAVISK EGILTGSHWG AIRATVKDGR FVAAKPFELD KYPSKMIAGL PDHVHNAARI RYPMVRVDWL RKRHLSDTSQ RGDNRFVRVS WDEALDMFYE ELERVQKTHG PSALLTASGW QSTGMFHNAS GMLAKAIALH GNSVGTGGDY STGAAQVILP RVVGSMEVYE QQTSWPLVLQ NSKTIVLWGS DLLKNQQANW WCPDHDVYEY YAQLKAKVAA GEIEVISIDP VVTSTHEYLG REHVKHIAVN PQTDVPLQLA LAHTLYSENL YDKNFLANYC VGFEQFLPYL LGEKDGQPKD AAWAEKLTGI DAETIRGLAR QMAANRTQII AGWCVQRMQH GEQWAWMIVV LAAMLGQIGL PGGGFGFGWH YNGAGTPGRK GVILSGFSGS TSIPPVHDNS DYKGYSSTIP IARFIDAILE PGKVINWNGK SVKLPPLKMC IFAGTNPFHR HQQINRIIEG LRKLETVIAI DNQWTSTCRF ADIVLPATTQ FERNDLDQYG NHSNRGIIAM KQVVPPQFEA RNDFDIFREL CRRFNREEAF TEGLDEMGWL KRIWQEGVQQ GKGRGVHLPA FDDFWNNKEY VEFDHPQMFV RHQAFREDPD LEPLGTPSGL IEIYSKTIAD MNYDDCQGHP MWFEKIERSH GGPGSQKYPL HLQSVHPDFR LHSQLCESET LRQQYTVAGK EPVFINPQDA SARGIRNGDV VRVFNARGQV LAGAVVSDRY APGVARIHEG AWYDPDKGGE PGALCKYGNP NVLTIDIGTS QLAQATSAHT TLVEIEKYNG TVEQVTAFNG PVEMVAQCEY VPASQVKS |
| Length |
848 |
| Mol. Wt |
94.455 kDa |
| pI |
6.8 (calculated) |
| Extinction coefficient |
150,230 - 151,480 (calc based on 27 Y, 20 W, and 10 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0003372 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG11814 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11814 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120001747 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1761 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
Ecoli K-12 |
0.811+/-0.273 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.001767478 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
2a |
molecules/cell/generation |
|
Ribosome Profiling |
Low confidence in the sequencing data set. |
PMID: 24766808 |
|
Protein |
E. coli K-12 MG1655 |
3 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
0a |
molecules/cell/generation |
|
Ribosome Profiling |
Low confidence in the sequencing data set. |
PMID: 24766808 |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
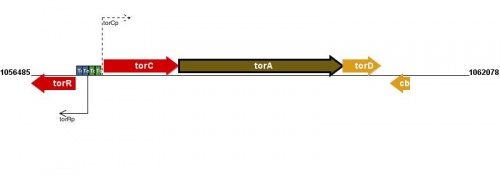 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:1058459..1058499
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for torA | |
|
microarray |
Summary of data for torA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to torA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11814 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1761 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11814 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001747 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0003372 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
<protect>
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
| edit table |
</protect>
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11814 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11814 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001747 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1761 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0003372 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories


