thiP:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
thiP |
|---|---|
| Gene Synonym(s) |
ECK0068, b0067, JW0066, sfuB, yabK[1], yabK |
| Product Desc. |
Component of thiamin ABC transporter[2][3] Thiamine and thiamine pyrophosphate ABC transporter[4] |
| Product Synonyms(s) |
fused thiamin transporter subunits of ABC superfamily: membrane components[1], B0067[2][1], YabK[2][1], SfuB[2][1], ThiP[2][1] , ECK0068, JW0066, sfuB, yabK, b0067 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Primarily characterized in Salmonella[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
thiP |
|---|---|
| Mnemonic |
Thiamin (and thiazole) |
| Synonyms |
ECK0068, b0067, JW0066, sfuB, yabK[1], yabK |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
1.57 minutes |
MG1655: 74521..72911 |
||
|
NC_012967: 77325..75715 |
||||
|
NC_012759: 72910..74520 |
||||
|
W3110 |
|
W3110: 74521..72911 |
||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
yabK::Tn5KAN-I-SceI (FB20007) |
Insertion at nt 1064 in Plus orientation |
PMID:15262929 |
contains pKD46 | ||||
|
yabK::Tn5KAN-I-SceI (FB20008) |
Insertion at nt 1064 in Plus orientation |
PMID:15262929 |
does not contain pKD46 | ||||
|
thiP(del) (Keio:JW0066) |
deletion |
deletion |
PMID:16738554 |
||||
|
thiP774(del)::kan |
PMID:16738554 |
||||||
|
Strain BW2952 |
T18C C64T A91G T205G G211A C352G T268C C283G T298C G304A C344T T409C G418A A421G A503G C535A G802T A834C G843A T880C G893A G907T C943T C958G C1009T TA1074-1075CG T1099C C1005T G1117A C1159T C1171T A1174G G1177C G1295A A1300G C1312T A1324G T1398C T1411C |
PMID:19376874 |
|||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW0066 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCGCAACGCGCCGTCAGCCGTT Primer 2:CCGTCAGTTTTAACATTTCGCCC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 18% [6] | ||
|
leuO3051::Tn10 |
Linked marker |
est. P1 cotransduction: 69% [6] | |
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11573 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11573 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001525 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB1533 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000246 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
ThiP |
|---|---|
| Synonyms |
fused thiamin transporter subunits of ABC superfamily: membrane components[1], B0067[2][1], YabK[2][1], SfuB[2][1], ThiP[2][1] , ECK0068, JW0066, sfuB, yabK, b0067 |
| Product description |
Component of thiamin ABC transporter[2][3] Thiamine and thiamine pyrophosphate ABC transporter[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of thiamin ABC transporter |
could be indirect |
||
|
Protein |
ydbK |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
plasma membrane |
From EcoCyc[3] |
| ||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MATRRQPLIP GWLIPGVSAT TLVVAVALAA FLALWWNAPQ DDWVAVWQDS YLWHVVRFSF WQAFLSALLS VIPAIFLARA LYRRRFPGRL ALLRLCAMTL ILPVLVAVFG ILSVYGRQGW LATLCQSLGL EWTFSPYGLQ GILLAHVFFN LPMASRLLLQ ALENIPGEQR QLAAQLGMRS WHFFRFVEWP WLRRQIPPVA ALIFMLCFAS FATVLSLGGG PQATTIELAI YQALSYDYDP ARAAMLALLQ MVCCLGLVLL SQRLSKAIAP GTTLLQGWRD PDDRLHSRIC DTVLIVLALL LLLPPLLAVI VDGVNRQLPE VLAQPVLWQA LWTSLRIALA AGVLCVVLTM MLLWSSRELR ARQKMLAGQV LEMSGMLILA MPGIVLATGF FLLLNNTIGL PQSADGIVIF TNALMAIPYA LKVLENPMRD ITARYSMLCQ SLGIEGWSRL KVVELRALKR PLAQALAFAC VLSIGDFGVV ALFGNDDFRT LPFYLYQQIG SYRSQDGAVT ALILLLLCFL LFTVIEKLPG RNVKTD |
| Length |
536 |
| Mol. Wt |
59.535 kDa |
| pI |
9.5 (calculated) |
| Extinction coefficient |
111,380 - 112,630 (calc based on 12 Y, 17 W, and 10 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0000246 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG11573 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11573 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120001525 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1533 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
34 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
41 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
10a |
molecules/cell/generation |
|
Ribosome Profiling |
Low confidence in the sequencing data set. |
PMID: 24766808 |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
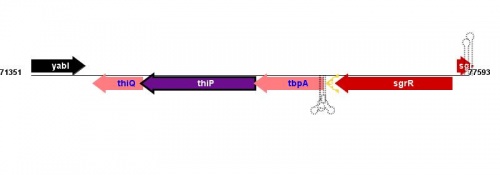 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:74501..74541
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for thiP | |
|
microarray |
Summary of data for thiP from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to thiP Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11573 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1533 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11573 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001525 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000246 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
YABK |
From SHIGELLACYC |
|
E. coli O157 |
YABK |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
PF00528 Binding-protein-dependent transport system inner membrane component |
||
|
PF00528 Binding-protein-dependent transport system inner membrane component |
||
|
EcoCyc:EG11573 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11573 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001525 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1533 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000246 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 3.4 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories



