secA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
secA |
|---|---|
| Gene Synonym(s) |
ECK0099, b0098, JW0096, azi, pea, prlD[1], prlD |
| Product Desc. |
Component of Sec Protein Secretion Complex[2][3] Translocon ATPase; targets protein precursors to the SecYE core translocon for secretion; autogenous translational repressor; ATP-dependent helicase activity on secMA mRNA; homodimeric/monomeric[4] |
| Product Synonyms(s) |
preprotein translocase subunit, ATPase[1], B0098[2][1], Pea[2][1], Azi[2][1], PrlD[2][1], SecA[2][1] , azi, ECK0099, JW0096, pea, prlD, b0098 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression |
transcription unit(s): secMA-mutT[2] |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Helicase activity is dispensable for both functions. Oligomerization induced by signal peptides. secA is regulated by the secretion competence of the SecM signal peptide. HT_Cmplx33_Cyt: AcpP+PurB+SecA. HT_Cmplx48_Mem: SecA+YdgA+YjiY.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
secA |
|---|---|
| Mnemonic |
Secretory |
| Synonyms |
ECK0099, b0098, JW0096, azi, pea, prlD[1], prlD |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
2.33 minutes |
MG1655: 108279..110984 |
||
|
NC_012967: 111083..113788 |
||||
|
NC_012759: 108278..110983 |
||||
|
W3110 |
|
W3110: 108279..110984 |
||
|
DH10B: 82383..85088 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
108279 |
Edman degradation |
PMID:2841285 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
secA207(aziR) |
azide resistant |
||||||
|
secA206(aziR) |
azide resistant |
||||||
|
secA214(aziR) |
azide resistant |
||||||
|
secA215 |
|||||||
|
secA204(aziR) |
azide resistant |
||||||
|
secA209(aziR) |
azide resistant |
||||||
|
secA216 |
|||||||
|
secA201(aziR) |
azide resistant |
||||||
|
secA202(aziR) |
azide resistant |
||||||
|
secA203(aziR) |
azide resistant |
||||||
|
secA205(aziR) |
azide resistant |
||||||
|
secA51(ts) |
temperature sensitive |
PMID:7665473 |
| ||||
| edit table |
<protect></protect>
Notes
SecA mutants were first identified by Oliver and Beckwith[5] by looking for suppressors of the Lac- phenotype of a maE-lacZ fusion. The original secA alleles were ts lethals.
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW0096 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCCTAATCAAATTGTTAACTAA Primer 2:CCTTGCAGGCGGCCATGGCACTG | |
|
Kohara Phage |
PMID:3038334 | ||
|
leuO3051::Tn10 |
Linked marker |
est. P1 cotransduction: 39% [7] | |
|
Linked marker |
est. P1 cotransduction: 18% [7] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10936 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10936 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000925 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0929 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000343 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
SecA |
|---|---|
| Synonyms |
preprotein translocase subunit, ATPase[1], B0098[2][1], Pea[2][1], Azi[2][1], PrlD[2][1], SecA[2][1] , azi, ECK0099, JW0096, pea, prlD, b0098 |
| Product description |
Component of Sec Protein Secretion Complex[2][3] Translocon ATPase; targets protein precursors to the SecYE core translocon for secretion; autogenous translational repressor; ATP-dependent helicase activity on secMA mRNA; homodimeric/monomeric[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005515 |
protein binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011130 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005515 |
protein binding |
PMID:17229438 |
IPI: Inferred from Physical Interaction |
F |
Seeded from EcoCyc (v14.0) |
Missing: with/from | ||
|
GO:0005515 |
protein binding |
PMID:17229438 |
IPI: Inferred from Physical Interaction |
EcoliWiki:secA |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_01382 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000185 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011115 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0963 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0086 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
PMID:1837021 |
IDA: Inferred from Direct Assay |
C |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0997 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-1003 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005886 |
plasma membrane |
PMID:1837021 |
IDA: Inferred from Direct Assay |
C |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0006605 |
protein targeting |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_01382 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006605 |
protein targeting |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000185 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008270 |
zinc ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0862 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0017038 |
protein import |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011115 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0017038 |
protein import |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011116 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005515 |
protein binding |
PMID:17229438 |
IPI: Inferred from Physical Interaction |
EcoliWiki:secA |
F |
E. coli SecA is a homodimer. |
complete | |
|
GO:0017038 |
protein import |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011130 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000287 |
magnesium ion binding |
PMID:17229438 |
IPI: Inferred from Physical Interaction |
F |
Missing: with/from | |||
|
GO:0065002 |
intracellular protein transmembrane transport |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_01382 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0065002 |
intracellular protein transmembrane transport |
PMID:6279567 |
IMP: Inferred from Mutant Phenotype |
P |
secA51(Ts) |
complete | ||
|
GO:0065002 |
intracellular protein transmembrane transport |
PMID:6279567 |
IMP: Inferred from Mutant Phenotype |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0015031 |
protein transport |
PMID:6279567 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0006605 |
protein targeting |
PMID:6279567 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0005737 |
cytoplasm |
PMID:1837021 |
IDA: Inferred from Direct Assay |
C |
Dimeric form of SecA is cytoplasmically localized (Figure 2 & 4). |
complete | ||
|
GO:0005886 |
plasma membrane |
PMID:1837021 |
IDA: Inferred from Direct Assay |
C |
SecA is also strongly associated with the plasma membrane (Figure 5). |
complete | ||
|
GO:0043952 |
protein transport by the Sec complex |
PMID:6279567 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of Sec Protein Secretion Complex |
could be indirect |
||
|
Protein |
ycfB |
PMID:15690043 |
Experiment(s):EBI-886105 | |
|
Protein |
ycaO |
PMID:15690043 |
Experiment(s):EBI-880477 | |
|
Protein |
dnaK |
PMID:15690043 |
Experiment(s):EBI-880477 | |
|
Protein |
gatZ |
PMID:15690043 |
Experiment(s):EBI-880477 | |
|
Protein |
infC |
PMID:15690043 |
Experiment(s):EBI-880477 | |
|
Protein |
minD |
PMID:15690043 |
Experiment(s):EBI-880477 | |
|
Protein |
rho |
PMID:15690043 |
Experiment(s):EBI-880477 | |
|
Protein |
secB |
PMID:15690043 |
Experiment(s):EBI-880477, EBI-880504, EBI-886105 | |
|
Protein |
tufA |
PMID:15690043 |
Experiment(s):EBI-880477, EBI-889981, EBI-886105, EBI-894882 | |
|
Protein |
rplL |
PMID:15690043 |
Experiment(s):EBI-886105 | |
|
Protein |
rplV |
PMID:15690043 |
Experiment(s):EBI-886105 | |
|
Protein |
rpsB |
PMID:15690043 |
Experiment(s):EBI-886105 | |
|
Protein |
ybiU |
PMID:15690043 |
Experiment(s):EBI-886105 | |
|
Protein |
aas |
PMID:15690043 |
Experiment(s):EBI-886105 | |
|
Protein |
norV |
PMID:15690043 |
Experiment(s):EBI-886105 | |
|
Protein |
bioB |
PMID:15690043 |
Experiment(s):EBI-886105 | |
|
Protein |
motA |
PMID:15690043 |
Experiment(s):EBI-886105 | |
|
Protein |
rplK |
PMID:15690043 |
Experiment(s):EBI-886105 | |
|
Protein |
acpP |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):18.837554 | |
|
Protein |
fimA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplK |
PMID:19402753 |
LCMS(ID Probability):99.4 | |
|
Protein |
rpsP |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
hns |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):14.030777 | |
|
Protein |
secB |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):17.040815 | |
|
Protein |
rpsJ |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
cspE |
PMID:19402753 |
LCMS(ID Probability):98.9 MALDI(Z-score):17.462556 | |
|
Protein |
rpmB |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
kefA |
PMID:19402753 |
MALDI(Z-score):18.451503 | |
|
Protein |
rho |
PMID:19402753 |
MALDI(Z-score):34.875526 | |
|
Protein |
metK |
PMID:19402753 |
MALDI(Z-score):21.897250 | |
|
Protein |
mnmA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplD |
PMID:19402753 |
LCMS(ID Probability):99.4 MALDI(Z-score):17.053160 | |
|
Protein |
minD |
PMID:19402753 |
MALDI(Z-score):30.779902 | |
|
Protein |
norV |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
gatZ |
PMID:19402753 |
MALDI(Z-score):21.103314 | |
|
Protein |
aas |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
motA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
bioB |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
ybiU |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplL |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MLIKLLTKVF GSRNDRTLRR MRKVVNIINA MEPEMEKLSD EELKGKTAEF RARLEKGEVL ENLIPEAFAV VREASKRVFG MRHFDVQLLG GMVLNERCIA EMRTGEGKTL TATLPAYLNA LTGKGVHVVT VNDYLAQRDA ENNRPLFEFL GLTVGINLPG MPAPAKREAY AADITYGTNN EYGFDYLRDN MAFSPEERVQ RKLHYALVDE VDSILIDEAR TPLIISGPAE DSSEMYKRVN KIIPHLIRQE KEDSETFQGE GHFSVDEKSR QVNLTERGLV LIEELLVKEG IMDEGESLYS PANIMLMHHV TAALRAHALF TRDVDYIVKD GEVIIVDEHT GRTMQGRRWS DGLHQAVEAK EGVQIQNENQ TLASITFQNY FRLYEKLAGM TGTADTEAFE FSSIYKLDTV VVPTNRPMIR KDLPDLVYMT EAEKIQAIIE DIKERTAKGQ PVLVGTISIE KSELVSNELT KAGIKHNVLN AKFHANEAAI VAQAGYPAAV TIATNMAGRG TDIVLGGSWQ AEVAALENPT AEQIEKIKAD WQVRHDAVLE AGGLHIIGTE RHESRRIDNQ LRGRSGRQGD AGSSRFYLSM EDALMRIFAS DRVSGMMRKL GMKPGEAIEH PWVTKAIANA QRKVESRNFD IRKQLLEYDD VANDQRRAIY SQRNELLDVS DVSETINSIR EDVFKATIDA YIPPQSLEEM WDIPGLQERL KNDFDLDLPI AEWLDKEPEL HEETLRERIL AQSIEVYQRK EEVVGAEMMR HFEKGVMLQT LDSLWKEHLA AMDYLRQGIH LRGYAQKDPK QEYKRESFSM FAAMLESLKY EVISTLSKVQ VRMPEEVEEL EQQRRMEAER LAQMQQLSHQ DDDSAAAAAL AAQTGERKVG RNDPCPCGSG KKYKQCHGRL Q |
| Length |
901 |
| Mol. Wt |
102.024 kDa |
| pI |
5.4 (calculated) |
| Extinction coefficient |
75,750 - 76,250 (calc based on 25 Y, 7 W, and 4 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0000343 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10936 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10936 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000925 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0929 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
5.18E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
71.319+/-0.353 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.104805915 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
3987 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
1148 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
3269 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
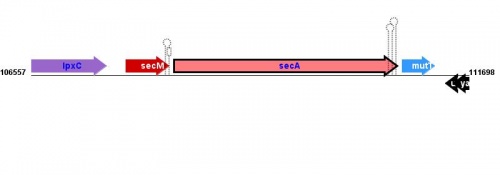 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:108259..108299
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for secA | |
|
microarray |
Summary of data for secA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (108145..108338) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to secA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10936 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0929 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10936 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000925 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000343 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Tetraodon nigroviridis |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
SECA |
From SHIGELLACYC |
|
E. coli O157 |
SECA |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10936 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10936 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000925 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0929 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000343 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ Oliver, DB & Beckwith, J (1981) E. coli mutant pleiotropically defective in the export of secreted proteins. Cell 25 765-72 PubMed
- ↑ 6.0 6.1 CGSC: The Coli Genetics Stock Center
- ↑ 7.0 7.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories
- Genes in OpenBioSystems with Promoter Fusions
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Oryza gramene
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Tetraodon nigroviridis
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


