recC:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
recC |
|---|---|
| Gene Synonym(s) |
ECK2818, b2822, JW2790[1], JW2790 |
| Product Desc. |
DNA helicase, ATP-dependent dsDNA/ssDNA exonuclease V subunit, ssDNA endonuclease[2][3]; RecBCD Exonuclease V subunit, recombination and repair; chi-activated RecBCD recombinase subunit; binds RecB and RecD; may confer processivity on holoenzyme[4] |
| Product Synonyms(s) |
exonuclease V (RecBCD complex), gamma chain[1], B2822[2][1], RecC[2][1] , ECK2818, JW2790, b2822 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
RecBCD is a bipolar helicase.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
recC |
|---|---|
| Mnemonic |
Recombination |
| Synonyms |
ECK2818, b2822, JW2790[1], JW2790 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
63.73 minutes |
MG1655: 2960450..2957082 |
||
|
NC_012967: 2857047..2853679 |
||||
|
NC_012759: 2844230..2847598 |
||||
|
W3110 |
|
W3110: 2961084..2957716 |
||
|
DH10B: 3054320..3050952 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
2957082 |
Edman degradation |
PMID:3520484 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
recC(del) (Keio:JW2790) |
deletion |
deletion |
PMID:16738554 |
||||
|
recC22 |
PMID:350827 |
||||||
|
recC1010(Nuc-) |
ds DNA exonuclease-minus |
||||||
|
recC73 |
PMID:350827 |
||||||
|
recC1002(Chi-) |
|||||||
|
recC1003(Chi-) |
|||||||
|
recC1004(Chi-) |
|||||||
|
recC1001(Chi-) |
|||||||
|
recC23 |
|||||||
|
recC |
deletion |
Sensitivity to |
decreases sensitivity to Bicyclomycin |
PMID:21357484 |
fig 2 | ||
|
SMR6039 recC::Tn10DCam |
Insertion |
SOS-Induction |
Decrease in spontaneous SOS-Induction |
PMID:23224554 |
Parent Strain: SMR11941 Experimental Strain: SMR17043 |
The mutation conferred a decrease in spontaneous SOS-induction. See table S7 for a summary of results. | |
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW2790 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCTTAAGGGTCTACCATTCCAA Primer 2:CCTGACTGATTAAAGCGAAACAG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 81% [6] | ||
|
Linked marker |
est. P1 cotransduction: % [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10825 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10825 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000816 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0818 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0009255 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
RecC |
|---|---|
| Synonyms |
exonuclease V (RecBCD complex), gamma chain[1], B2822[2][1], RecC[2][1] , ECK2818, JW2790, b2822 |
| Product description |
DNA helicase, ATP-dependent dsDNA/ssDNA exonuclease V subunit, ssDNA endonuclease[2][3]; RecBCD Exonuclease V subunit, recombination and repair; chi-activated RecBCD recombinase subunit; binds RecB and RecD; may confer processivity on holoenzyme[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0005737 |
cytoplasm |
C |
Seeded from Riley et al 2006 [1]. |
Missing: evidence, reference | ||||
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004386 |
helicase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0347 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004518 |
nuclease activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0540 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004519 |
endonuclease activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0255 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004527 |
exonuclease activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0269 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006281 |
DNA repair |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0234 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006974 |
response to DNA damage stimulus |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0227 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008854 |
exodeoxyribonuclease V activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006697 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008854 |
exodeoxyribonuclease V activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:3.1.11.5 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009338 |
exodeoxyribonuclease V complex |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006697 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016787 |
hydrolase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0378 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of recBCD |
could be indirect |
||
|
Protein |
uvrA |
PMID:16606699 |
Experiment(s):EBI-1144074 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MLRVYHSNRL DVLEALMEFI VERERLDDPF EPEMILVQST GMAQWLQMTL SQKFGIAANI DFPLPASFIW DMFVRVLPEI PKESAFNKQS MSWKLMTLLP QLLEREDFTL LRHYLTDDSD KRKLFQLSSK AADLFDQYLV YRPDWLAQWE TGHLVEGLGE AQAWQAPLWK ALVEYTHQLG QPRWHRANLY QRFIETLESA TTCPPGLPSR VFICGISALP PVYLQALQAL GKHIEIHLLF TNPCRYYWGD IKDPAYLAKL LTRQRRHSFE DRELPLFRDS ENAGQLFNSD GEQDVGNPLL ASWGKLGRDY IYLLSDLESS QELDAFVDVT PDNLLHNIQS DILELENRAV AGVNIEEFSR SDNKRPLDPL DSSITFHVCH SPQREVEVLH DRLLAMLEED PTLTPRDIIV MVADIDSYSP FIQAVFGSAP ADRYLPYAIS DRRARQSHPV LEAFISLLSL PDSRFVSEDV LALLDVPVLA ARFDITEEGL RYLRQWVNES GIRWGIDDDN VRELELPATG QHTWRFGLTR MLLGYAMESA QGEWQSVLPY DESSGLIAEL VGHLASLLMQ LNIWRRGLAQ ERPLEEWLPV CRDMLNAFFL PDAETEAAMT LIEQQWQAII AEGLGAQYGD AVPLSLLRDE LAQRLDQERI SQRFLAGPVN ICTLMPMRSI PFKVVCLLGM NDGVYPRQLA PLGFDLMSQK PKRGDRSRRD DDRYLFLEAL ISAQQKLYIS YIGRSIQDNS ERFPSVLVQE LIDYIGQSHY LPGDEALNCD ESEARVKAHL TCLHTRMPFD PQNYQPGERQ SYAREWLPAA SQAGKAHSEF VQPLPFTLPE TVPLETLQRF WAHPVRAFFQ MRLQVNFRTE DSEIPDTEPF ILEGLSRYQI NQQLLNALVE QDDAERLFRR FRAAGDLPYG AFGEIFWETQ CQEMQQLADR VIACRQPGQS MEIDLACNGV QITGWLPQVQ PDGLLRWRPS LLSVAQGMQL WLEHLVYCAS GGNGESRLFL RKDGEWRFPP LAAEQALHYL SQLIEGYREG MSAPLLVLPE SGGAWLKTCY DAQNDAMLDD DSTLQKARTK FLQAYEGNMM VRGEGDDIWY QRLWRQLTPE TMEAIVEQSQ RFLLPLFRFN QS |
| Length |
1,122 |
| Mol. Wt |
128.851 kDa |
| pI |
4.7 (calculated) |
| Extinction coefficient |
200,650 - 202,400 (calc based on 35 Y, 27 W, and 14 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0009255 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10825 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10825 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000816 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0818 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
118 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
41 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
88 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
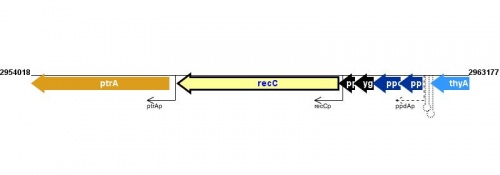 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:2960430..2960470
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for recC | |
|
microarray |
Summary of data for recC from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to recC Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10825 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0818 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10825 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000816 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0009255 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Shigella flexneri |
RECC |
From SHIGELLACYC |
|
E. coli O157 |
RECC |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10825 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10825 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000816 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0818 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0009255 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories


