putA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
putA |
|---|---|
| Gene Synonym(s) |
ECK1005, b1014, JW0999, poaA, putC, proline utilization A[1][2] |
| Product Desc. |
PutA bifunctional enzyme and transcriptional regulator[2]; PutA transcriptional repressor, Proline dehydrogenase/pyrroline-5-carboxylate dehydrogenase[3]; Component of PUTA-CPLX[2]; PutA transcriptional repressor, Proline dehydrogenase/pyrroline-5-carboxylate dehydrogenase[3] Proline dehydrogenase and repressor for the putAP divergon[4] |
| Product Synonyms(s) |
fused DNA-binding transcriptional regulator[1], proline dehydrogenase[1], pyrroline-5-carboxylate dehydrogenase[1], B1014[2][1], PutC[2][1], PoaA[2][1], PutA[2][1] , ECK1005, JW0999, poaA, putC, b1014 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
PutA is a multifunctional membrane-associated flavoprotein involved in proline catabolism: the central proline dehydrogenese domain feeds its product to the C-terminal Delta(1)-pyrroline-5-carboxylate dehydrogenese domain, to produce glutamate. The N-terminal 90 residue domain is the DNA-binding repressor domain. A divergon is a regulon comprised of two adjacent divergent operons or genes. Binds TrxA (Kumar, 2004).[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
putA |
|---|---|
| Mnemonic |
Proline utilization |
| Synonyms |
ECK1005, b1014, JW0999, poaA, putC, proline utilization A[1][2] |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
23.15 minutes |
MG1655: 1078105..1074143 |
||
|
NC_012967: 1095166..1091204 |
||||
|
NC_012759: 977111..981073 |
||||
|
W3110 |
|
W3110: 1079304..1075342 |
||
|
DH10B: 1132033..1128071 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
putA(del) (Keio:JW0999) |
deletion |
deletion |
PMID:16738554 |
||||
|
putA11 |
|||||||
|
putA1::Tn5 |
PMID:7016835 |
||||||
|
put(del)A758::kan |
PMID:16738554 |
||||||
|
putAP(del)101 |
Auxotrophies |
Proline Auxotroph |
PMID:6355059 |
Strains: RM2 |
|||
|
putP putA + |
Resistant to |
Dehydroproline resistance |
PMID:7016835 |
Strain:CSH4 |
table 2 | ||
|
putP putA + |
Resistant to |
L-Azetidine-2-carboxylate resistance |
PMID:7016835 |
Strain:CSH4 |
table 2 | ||
|
putP in strain C4 |
Resistant to |
Dehydroproline resistance |
PMID:4865475 |
Strain: C4 |
Table 1 | ||
|
putP in strain C4 |
Resistant to |
L-Azetidine-2-carboxylate resistance |
PMID:4865475 |
Strain: C4 |
Table 1 | ||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW0999 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:gccGGAACCACCACCATGGGGGTTAAGCTG Primer 2:ccACCTATAGTCATTAAGCTGGCGTTACCG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 67% [6] | ||
|
Linked marker |
est. P1 cotransduction: % [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10801 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10801 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000792 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0794 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0003424 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
PutA |
|---|---|
| Synonyms |
fused DNA-binding transcriptional regulator[1], proline dehydrogenase[1], pyrroline-5-carboxylate dehydrogenase[1], B1014[2][1], PutC[2][1], PoaA[2][1], PutA[2][1] , ECK1005, JW0999, poaA, putC, b1014 |
| Product description |
PutA bifunctional enzyme and transcriptional regulator[2]; PutA transcriptional repressor, Proline dehydrogenase/pyrroline-5-carboxylate dehydrogenase[3]; Component of PUTA-CPLX[2]; PutA transcriptional repressor, Proline dehydrogenase/pyrroline-5-carboxylate dehydrogenase[3] Proline dehydrogenase and repressor for the putAP divergon[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0003677 |
DNA binding |
PMID:1618807 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0003700 |
transcription factor activity |
PMID:18586269 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0003842 |
1-pyrroline-5-carboxylate dehydrogenase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005933 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003842 |
1-pyrroline-5-carboxylate dehydrogenase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:1.5.1.12 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003842 |
1-pyrroline-5-carboxylate dehydrogenase activity |
PMID:1618807 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0004657 |
proline dehydrogenase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002872 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004657 |
proline dehydrogenase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:1.5.99.8 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004657 |
proline dehydrogenase activity |
PMID:1618807 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0005515 |
protein binding |
PMID:16606699 |
IPI: Inferred from Physical Interaction |
UniProtKB:P63389 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006350 |
transcription |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0804 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006537 |
glutamate biosynthetic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002872 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006561 |
proline biosynthetic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005933 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0010133 |
proline catabolic process to glutamate |
PMID:7006756 |
IMP: Inferred from Mutant Phenotype |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0010843 |
promoter binding |
PMID:18586269 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0016566 |
specific transcriptional repressor activity |
PMID:18586269 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0050660 |
FAD binding |
PMID:1618807 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0055114 |
oxidation reduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002872 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005933 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR015590 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR016160 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR016161 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR016162 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0560 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of PUTA-CPLX |
could be indirect |
||
|
Protein |
ydaC |
PMID:16606699 |
Experiment(s):EBI-1138642 | |
|
Protein |
eutM |
PMID:16606699 |
Experiment(s):EBI-1138642 | |
|
Protein |
yfaS |
PMID:16606699 |
Experiment(s):EBI-1138642 | |
|
Protein |
dnaK |
PMID:16606699 |
Experiment(s):EBI-1138642 | |
|
Protein |
mukF |
PMID:16606699 |
Experiment(s):EBI-1138642 | |
|
Protein |
groL |
PMID:16606699 |
Experiment(s):EBI-1138642 | |
|
Protein |
rplQ |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rpmB |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplP |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
yfiD |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplR |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rpsA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
ahpC |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
groS |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):2.778583 | |
|
Protein |
dnaN |
PMID:19402753 |
MALDI(Z-score):22.046630 | |
|
Protein |
elaB |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rpmC |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
hns |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
eno |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rplA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rpmG |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rpsP |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
hupA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
tufB |
PMID:19402753 |
MALDI(Z-score):18.820564 | |
|
Protein |
adk |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
phoA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
pgk |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):5.720428 | |
|
Protein |
rplI |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
fbaA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
hslV |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
ribE |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
cspC |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
talB |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
leuD |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
plasma membrane |
| |||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MGTTTMGVKL DDATRERIKS AATRIDRTPH WLIKQAIFSY LEQLENSDTL PELPALLSGA ANESDEAPTP AEEPHQPFLD FAEQILPQSV SRAAITAAYR RPETEAVSML LEQARLPQPV AEQAHKLAYQ LADKLRNQKN ASGRAGMVQG LLQEFSLSSQ EGVALMCLAE ALLRIPDKAT RDALIRDKIS NGNWQSHIGR SPSLFVNAAT WGLLFTGKLV STHNEASLSR SLNRIIGKSG EPLIRKGVDM AMRLMGEQFV TGETIAEALA NARKLEEKGF RYSYDMLGEA ALTAADAQAY MVSYQQAIHA IGKASNGRGI YEGPGISIKL SALHPRYSRA QYDRVMEELY PRLKSLTLLA RQYDIGINID AEESDRLEIS LDLLEKLCFE PELAGWNGIG FVIQAYQKRC PLVIDYLIDL ATRSRRRLMI RLVKGAYWDS EIKRAQMDGL EGYPVYTRKV YTDVSYLACA KKLLAVPNLI YPQFATHNAH TLAAIYQLAG QNYYPGQYEF QCLHGMGEPL YEQVTGKVAD GKLNRPCRIY APVGTHETLL AYLVRRLLEN GANTSFVNRI ADTSLPLDEL VADPVTAVEK LAQQEGQTGL PHPKIPLPRD LYGHGRDNSA GLDLANEHRL ASLSSALLNS ALQKWQALPM LEQPVAAGEM SPVINPAEPK DIVGYVREAT PREVEQALES AVNNAPIWFA TPPAERAAIL HRAAVLMESQ MQQLIGILVR EAGKTFSNAI AEVREAVDFL HYYAGQVRDD FANETHRPLG PVVCISPWNF PLAIFTGQIA AALAAGNSVL AKPAEQTPLI AAQGIAILLE AGVPPGVVQL LPGRGETVGA QLTGDDRVRG VMFTGSTEVA TLLQRNIASR LDAQGRPIPL IAETGGMNAM IVDSSALTEQ VVVDVLASAF DSAGQRCSAL RVLCLQDEIA DHTLKMLRGA MAECRMGNPG RLTTDIGPVI DSEAKANIER HIQTMRSKGR PVFQAVRENS EDAREWQSGT FVAPTLIELD DFAELQKEVF GPVLHVVRYN RNQLPELIEQ INASGYGLTL GVHTRIDETI AQVTGSAHVG NLYVNRNMVG AVVGVQPFGG EGLSGTGPKA GGPLYLYRLL ANRPESALAV TLARQDAKYP VDAQLKAALT QPLNALREWA ANRPELQALC TQYGELAQAG TQRLLPGPTG ERNTWTLLPR ERVLCIADDE QDALTQLAAV LAVGSQVLWP DDALHRQLVK ALPSAVSERI QLAKAENITA QPFDAVIFHG DSDQLRALCE AVAARDGTIV SVQGFARGES NILLERLYIE RSLSVNTAAA GGNASLMTIG |
| Length |
1,320 |
| Mol. Wt |
143.817 kDa |
| pI |
5.9 (calculated) |
| Extinction coefficient |
124,110 - 125,735 (calc based on 39 Y, 12 W, and 13 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0003424 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10801 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10801 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000792 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0794 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
3.29E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
12.847+/-0.126 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.009464917 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
222 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
64 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
334 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
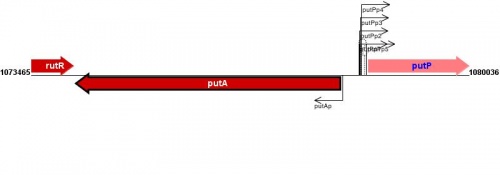 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:1078085..1078125
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for putA | |
|
microarray |
Summary of data for putA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to putA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10801 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0794 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10801 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000792 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0003424 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Drosophila pseudoobscura |
|
From Inparanoid:20070104 |
|
Saccharomyces cerevisiae |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
E. coli O157 |
PUTA |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10801 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10801 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000792 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0794 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0003424 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories


