pflB:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
pflB |
|---|---|
| Gene Synonym(s) |
ECK0894, b0903, JW0886, pfl[1], pfl |
| Product Desc. |
Component of pyruvate formate-lyase (inactive)[2][3] Pyruvate formate lyase I; induced anaerobically; radical activated by PflA; deactivated by AdhE[4] |
| Product Synonyms(s) |
pyruvate formate lyase I[1], B0903[2][1], Pfl[2][1], PflB[2][1] , ECK0894, JW0886, pfl, b0903 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
HT_Cmplx8_Cyt: YfiD+PflB.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
pflB |
|---|---|
| Mnemonic |
Pyruvate formate lyase |
| Synonyms |
ECK0894, b0903, JW0886, pfl[1], pfl |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
20.49 minutes |
MG1655: 952777..950495 |
||
|
NC_012967: 970647..968365 |
||||
|
NC_012759: 853463..855745 |
||||
|
W3110 |
|
W3110: 953976..951694 |
||
|
DH10B: 1006705..1004423 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
950498 |
Edman degradation |
PMID:3053170 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔpflB (Keio:JW0886) |
deletion |
deletion |
PMID:16738554 |
||||
|
pflB1 |
|||||||
|
Δpfl-2::Cm |
|||||||
|
ΔpflB727::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW0886 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCTCCGAGCTTAATGAAAAGTT Primer 2:CCCATAGATTGAGTGAAGGTACG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
zca-1230::Tn10 |
Linked marker |
est. P1 cotransduction: 88% [6] | |
|
Linked marker |
est. P1 cotransduction: 9% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10701 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10701 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000693 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0695 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0003071 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
PflB |
|---|---|
| Synonyms |
pyruvate formate lyase I[1], B0903[2][1], Pfl[2][1], PflB[2][1] , ECK0894, JW0886, pfl, b0903 |
| Product description |
Component of pyruvate formate-lyase (inactive)[2][3] Pyruvate formate lyase I; induced anaerobically; radical activated by PflA; deactivated by AdhE[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of pyruvate formate-lyase (inactive) |
could be indirect |
||
|
Protein |
tpiA |
PMID:15690043 |
Experiment(s):EBI-882596, EBI-879853, EBI-884726 | |
|
Protein |
grcA |
PMID:15690043 |
Experiment(s):EBI-882596, EBI-884695, EBI-891531, EBI-890164 | |
|
Protein |
tpiA |
PMID:16606699 |
Experiment(s):EBI-1138311 | |
|
Protein |
nadE |
PMID:16606699 |
Experiment(s):EBI-1138311 | |
|
Protein |
matC |
PMID:16606699 |
Experiment(s):EBI-1138311 | |
|
Protein |
fixB |
PMID:16606699 |
Experiment(s):EBI-1138311 | |
|
Protein |
ydfK |
PMID:16606699 |
Experiment(s):EBI-1138311 | |
|
Protein |
fhuA |
PMID:16606699 |
Experiment(s):EBI-1138311 | |
|
Protein |
zwf |
PMID:15690043 |
Experiment(s):EBI-890164 | |
|
Protein |
pykA |
PMID:15690043 |
Experiment(s):EBI-890164 | |
|
Protein |
pykA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
ygjD |
PMID:19402753 |
MALDI(Z-score):35.025327 | |
|
Protein |
zwf |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MSELNEKLAT AWEGFTKGDW QNEVNVRDFI QKNYTPYEGD ESFLAGATEA TTTLWDKVME GVKLENRTHA PVDFDTAVAS TITSHDAGYI NKQLEKIVGL QTEAPLKRAL IPFGGIKMIE GSCKAYNREL DPMIKKIFTE YRKTHNQGVF DVYTPDILRC RKSGVLTGLP DAYGRGRIIG DYRRVALYGI DYLMKDKLAQ FTSLQADLEN GVNLEQTIRL REEIAEQHRA LGQMKEMAAK YGYDISGPAT NAQEAIQWTY FGYLAAVKSQ NGAAMSFGRT STFLDVYIER DLKAGKITEQ EAQEMVDHLV MKLRMVRFLR TPEYDELFSG DPIWATESIG GMGLDGRTLV TKNSFRFLNT LYTMGPSPEP NMTILWSEKL PLNFKKFAAK VSIDTSSLQY ENDDLMRPDF NNDDYAIACC VSPMIVGKQM QFFGARANLA KTMLYAINGG VDEKLKMQVG PKSEPIKGDV LNYDEVMERM DHFMDWLAKQ YITALNIIHY MHDKYSYEAS LMALHDRDVI RTMACGIAGL SVAADSLSAI KYAKVKPIRD EDGLAIDFEI EGEYPQFGNN DPRVDDLAVD LVERFMKKIQ KLHTYRDAIP TQSVLTITSN VVYGKKTGNT PDGRRAGAPF GPGANPMHGR DQKGAVASLT SVAKLPFAYA KDGISYTFSI VPNALGKDDE VRKTNLAGLM DGYFHHEASI EGGQHLNVNV MNREMLLDAM ENPEKYPQLT IRVSGYAVRF NSLTKEQQQD VITRTFTQSM |
| Length |
760 |
| Mol. Wt |
85.359 kDa |
| pI |
5.9 (calculated) |
| Extinction coefficient |
89,160 - 89,785 (calc based on 34 Y, 7 W, and 5 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0003071 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10701 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10701 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000693 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0695 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
1.21E+04 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
238.944+/-1.657 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
1.962+/-0.071 |
Molecules/cell |
|
by FISH |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.388913234 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
10029 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
5982 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
6696 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
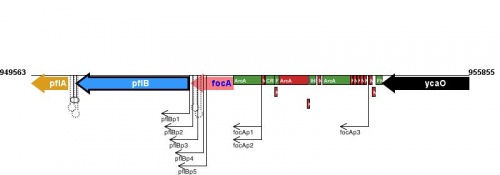 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:952757..952797
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for pflB | |
|
microarray |
Summary of data for pflB from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to pflB Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10701 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0695 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10701 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000693 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0003071 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Shigella flexneri |
PFLB |
From SHIGELLACYC |
|
E. coli O157 |
PFLB |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10701 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10701 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000693 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0695 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0003071 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories


