pepA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
pepA |
|---|---|
| Gene Synonym(s) |
ECK4253, b4260, JW4217, carP, xerB[1], xerB |
| Product Desc. |
PepA Transcriptional Repressor, Aminopeptidase A/I[2][3] Multifunctional Aminopeptidase A; transcriptional regulator of carAB; non-peptidase, DNA-binding role in site-specific recombination mediating ColE1 plasmid multimer resolution; homohexameric[4] |
| Product Synonyms(s) |
aminopeptidase A, a cyteinylglycinase[1], PepA[2][1], B4260[2][1], XerB[2][1], CarP[2][1] , carP, ECK4253, JW4217, xerB, b4260 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
PepA has a broad specificity for successive removal of many N-terminal amino acids, although leucine is preferred as the residue N-terminal to the cleavage site.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
pepA |
|---|---|
| Mnemonic |
Peptidase |
| Synonyms |
ECK4253, b4260, JW4217, carP, xerB[1], xerB |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
96.61 minutes, 96.61 minutes |
MG1655: 4483974..4482463 |
||
|
NC_012967: 4469178..4467667 |
||||
|
NC_012759: 4421198..4422709 |
||||
|
W3110 |
|
W3110: 4490631..4489120 |
||
|
DH10B: 4585782..4584271 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
4482463 |
Edman degradation |
PMID:2670557 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
pepAE354A |
E354A |
Loss of activity |
seeded from UniProt:P68767 | ||||
|
ΔpepA (Keio:JW4217) |
deletion |
deletion |
PMID:16738554 |
||||
|
ΔpepA733::kan |
PMID:16738554 |
||||||
|
carP6 |
TGG (W473) into a TAG (amber stop) |
confers partial constitutive expression to the carAB promoter P1 and is trans-dominant over the wt gene |
PMID:7616564 |
||||
|
xerB1 |
Tn5 insertion that affects the regulation of carAB expression |
PMID:7616564 |
| ||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW4217 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCGAGTTTAGTGTAAAAAGCGG Primer 2:CCtTCTTCGCCGTTAAACCCAGC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 21% [6] | ||
|
Linked marker |
est. P1 cotransduction: % [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10694 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10694 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000686 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0688 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0013953 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
PepA |
|---|---|
| Synonyms |
aminopeptidase A, a cyteinylglycinase[1], PepA[2][1], B4260[2][1], XerB[2][1], CarP[2][1] , carP, ECK4253, JW4217, xerB, b4260 |
| Product description |
PepA Transcriptional Repressor, Aminopeptidase A/I[2][3] Multifunctional Aminopeptidase A; transcriptional regulator of carAB; non-peptidase, DNA-binding role in site-specific recombination mediating ColE1 plasmid multimer resolution; homohexameric[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
The xerB synonym was given when mutant alleles were isolated that are defective in site-specific recombination at the plasmid ColE1 cer locus.[7][8]
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0004177 |
aminopeptidase activity |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00181 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004177 |
aminopeptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000819 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004177 |
aminopeptidase activity |
PMID:2670557 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0004177 |
aminopeptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008283 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004177 |
aminopeptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011356 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004177 |
aminopeptidase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0031 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005622 |
intracellular |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000819 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005622 |
intracellular |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008283 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00181 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011356 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006508 |
proteolysis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR000819 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006508 |
proteolysis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008283 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008233 |
peptidase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0645 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
PMID:7616564 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0008235 |
metalloexopeptidase activity |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00181 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008235 |
metalloexopeptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011356 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0001073 |
DNA binding transcription antitermination factor activity |
PMID:7616564 |
IMP: Inferred from Mutant Phenotype |
F |
regulation of the carAB operon replaced obsolete GO:0016564 |
complete | ||
|
GO:0016564 |
transcription repressor activity |
PMID:7616564 |
IMP: Inferred from Mutant Phenotype |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0016787 |
hydrolase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0378 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0043171 |
peptide catabolic process |
PMID:355237 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0042150 |
plasmid recombination |
PMID:2670557 |
IMP: Inferred from Mutant Phenotype |
P |
XerC, XerD, ArgR, and PepA, are required for site-specific recombination at the ColE1 cer locus in E. coli K-12 (see references in [9]). |
complete | ||
|
GO:0019538 |
protein metabolic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011356 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030145 |
manganese ion binding |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00181 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030145 |
manganese ion binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011356 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030145 |
manganese ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0464 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046872 |
metal ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0479 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006276 |
plasmid maintenance |
PMID:2670557 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
sucB |
PMID:16606699 |
Experiment(s):EBI-1147774 | |
|
Protein |
pyrB |
PMID:16606699 |
Experiment(s):EBI-1147774 | |
|
Protein |
lacZ |
PMID:16606699 |
Experiment(s):EBI-1147774 | |
|
Protein |
lysU |
PMID:19402753 |
MALDI(Z-score):28.486162 | |
|
Protein |
pepD |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):27.154431 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MEFSVKSGSP EKQRSACIVV GVFEPRRLSP IAEQLDKISD GYISALLRRG ELEGKPGQTL LLHHVPNVLS ERILLIGCGK ERELDERQYK QVIQKTINTL NDTGSMEAVC FLTELHVKGR NNYWKVRQAV ETAKETLYSF DQLKTNKSEP RRPLRKMVFN VPTRRELTSG ERAIQHGLAI AAGIKAAKDL GNMPPNICNA AYLASQARQL ADSYSKNVIT RVIGEQQMKE LGMHSYLAVG QGSQNESLMS VIEYKGNASE DARPIVLVGK GLTFDSGGIS IKPSEGMDEM KYDMCGAAAV YGVMRMVAEL QLPINVIGVL AGCENMPGGR AYRPGDVLTT MSGQTVEVLN TDAEGRLVLC DVLTYVERFE PEAVIDVATL TGACVIALGH HITGLMANHN PLAHELIAAS EQSGDRAWRL PLGDEYQEQL ESNFADMANI GGRPGGAITA GCFLSRFTRK YNWAHLDIAG TAWRSGKAKG ATGRPVALLA QFLLNRAGFN GEE |
| Length |
503 |
| Mol. Wt |
54.88 kDa |
| pI |
7.3 (calculated) |
| Extinction coefficient |
42,860 - 43,985 (calc based on 14 Y, 4 W, and 9 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0013953 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10694 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10694 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000686 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0688 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
5.77E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
E. coli K-12 MG1655 |
2548 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
1308 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
2563 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
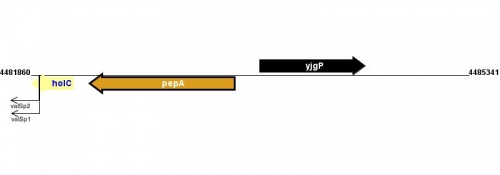 Figure courtesy of RegulonDB |
</protect>
Notes
Identification of the promoter region of carP and the regulation of its txn are discussed in reference [10]
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:4483954..4483994
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for pepA | |
|
microarray |
Summary of data for pepA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to pepA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10694 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0688 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10694 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000686 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0013953 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Danio rerio |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Drosophila melanogaster |
|
From Inparanoid:20070104 |
|
Drosophila pseudoobscura |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Macaca mulatta |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Tetraodon nigroviridis |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
PEPA |
From SHIGELLACYC |
|
E. coli O157 |
PEPA |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10694 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10694 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000686 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0688 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0013953 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Stirling, CJ et al. (1988) Multicopy plasmid stability in Escherichia coli requires host-encoded functions that lead to plasmid site-specific recombination. Mol. Gen. Genet. 214 80-4 PubMed
- ↑ Stirling, CJ et al. (1989) xerB, an Escherichia coli gene required for plasmid ColE1 site-specific recombination, is identical to pepA, encoding aminopeptidase A, a protein with substantial similarity to bovine lens leucine aminopeptidase. EMBO J. 8 1623-7 PubMed
- ↑ Sherratt, DJ et al. (1995) Site-specific recombination and circular chromosome segregation. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 347 37-42 PubMed
- ↑ Charlier, D et al. (1995) carP, involved in pyrimidine regulation of the Escherichia coli carbamoylphosphate synthetase operon encodes a sequence-specific DNA-binding protein identical to XerB and PepA, also required for resolution of ColEI multimers. J. Mol. Biol. 250 392-406 PubMed
Categories
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Apis mellifera
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Bos taurus
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Canis familiaris
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Danio rerio
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Drosophila melanogaster
- Genes with homologs in Drosophila pseudoobscura
- Genes with homologs in Gallus gallus
- Genes with homologs in Homo sapiens
- Genes with homologs in Macaca mulatta
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Oryza gramene
- Genes with homologs in Pan troglodytes
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Schizosaccharomyces pombe
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Tetraodon nigroviridis
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


