mutL:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
mutL |
|---|---|
| Gene Synonym(s) |
ECK4166, b4170, JW4128[1], JW4128 |
| Product Desc. |
Component of MutHLS complex, methyl-directed mismatch repair[2][3] Methyl-directed mismatch repair; matchmaker protein; weak DNA-stimulated ATPase[4] |
| Product Synonyms(s) |
methyl-directed mismatch repair protein[1], B4170[2][1], MutL[2][1] , ECK4166, JW4128, b4170 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression |
transcription unit(s): yjeFE-amiB-mutL-miaA-hfq-hflXKC[2], ORF55-ORF17-amiB-mutL-miaA-hfq-hflXKC |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
MutL DNA-binding is required for methyl-directed mismatch repair (Robertson, 2006). The ATPase activity of MutL is essential for mismatch repair (Ban, 1998; Ban, 1999; Spampinato, 2000; Robertson, 2006). MutL binds UvrD, stimulates UvrD unwinding activity and is required for UvrD loading onto DNA (Hall, 1998; Yamaguchi, 1998; Mechanic, 2000). MutL stimulates MutS and Vsr binding to mismatched DNA (Drotschmann, 1998).[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
mutL |
|---|---|
| Mnemonic |
Mutator |
| Synonyms |
ECK4166, b4170, JW4128[1], JW4128 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
94.74 minutes |
MG1655: 4395435..4397282 |
||
|
NC_012967: 4375567..4377414 |
||||
|
NC_012759: 4334170..4336017 |
||||
|
W3110 |
|
W3110: 4402092..4403939 |
||
|
DH10B: 4495797..4497644 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
mutL(del) (Keio:JW4128) |
deletion |
deletion |
PMID:16738554 |
||||
|
mutL218::Tn10 |
|||||||
|
mutL13 |
PMID:4919752 |
||||||
|
mutL720(del)::kan |
PMID:16738554 |
||||||
|
mutL::KmMlu |
GC To AT and AT to GC transition Mutation |
Mutation Frequency |
Insertion caused GC to AT and AT to GC transition mutation to increase. Table 2. |
PMID:1999389 |
Strain: NU1521 (AT to GC) Strain: NU1509 (GC to AT) | ||
|
mutL:omega(Kmr OR Cmr);BsaAI |
Auxotrophies |
The polar omega cassettes blocked mutL-miaA transcription and caused the full-lengthened protected species to disappear, Fig. 2 lanes 3 and 4. |
PMID:7748952 |
Strains: TX2568 (Cmr) & TX2569 (Kmr) | |||
|
MutL(del)335N |
deletion of N terminus |
excision activity |
Truncation mutant was inefficient as wild type MutL in promoting the excision activity of ExoX. Interaction between MutL and ExoX was also reduced. figure 4. These data suggest that the 335 N-terminal amoni acids of MutL are essential for both the interaction and stimulation of ExoX. |
PMID:20638361 |
|||
|
MutL-E29A |
replacement of Glutamic acid to Alanine at position 29. |
DNA repair |
mutant form of MutL is inactive in ATP hydrolysis but still has a physical interaction with RecA. Mutation had an inhibitory effect on RecA's ATPase activity, Figure 4B. |
PMID:22001225 |
Parent Strain:MG1655 |
| |
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW4128 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCCCAATTCAGGTCTTACCGCC Primer 2:CCtTCATCTTTCAGGGCTTTTAT | |
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 20% [6] | ||
|
Linked marker |
est. P1 cotransduction: 30% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11281 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11281 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001254 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB1258 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0013655 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
MutL |
|---|---|
| Synonyms |
methyl-directed mismatch repair protein[1], B4170[2][1], MutL[2][1] , ECK4166, JW4128, b4170 |
| Product description |
Component of MutHLS complex, methyl-directed mismatch repair[2][3] Methyl-directed mismatch repair; matchmaker protein; weak DNA-stimulated ATPase[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0006298 |
mismatch repair |
PMID:6987663 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0005524 |
ATP binding |
PMID:9827806 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0006298 |
mismatch repair |
PMID:2665076 |
IDA: Inferred from Direct Assay |
P |
complete | |||
|
GO:0032300 |
mismatch repair complex |
PMID:6987663 |
IGI: Inferred from Genetic Interaction |
UniProtKB:P06722|UniProtKB:P23909 |
C |
With MutH (P06722) and MutS (P23909). |
complete | |
|
GO:0005515 |
protein binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020667 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016887 |
ATPase activity |
PMID:9827806 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002099 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016887 |
ATPase activity |
PMID:9827806 |
IMP: Inferred from Mutant Phenotype |
F |
complete | |||
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003594 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013507 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006298 |
mismatch repair |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00149 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006298 |
mismatch repair |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002099 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006298 |
mismatch repair |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013507 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006298 |
mismatch repair |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR020667 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030983 |
mismatched DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002099 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030983 |
mismatched DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013507 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
PMID:12606120 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0003677 |
DNA binding |
PMID:12606120 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0016887 |
ATPase activity |
PMID:9827806 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0005524 |
ATP binding |
PMID:9827806 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0000018 |
regulation of DNA recombination |
PMID:2555716 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of MutHLS complex, methyl-directed mismatch repair |
PMID:6987663 |
Inferred from genetic interaction | |
|
Protein |
tufA |
PMID:15690043 |
Experiment(s):EBI-880976 | |
|
Protein |
dnaK |
PMID:15690043 |
Experiment(s):EBI-887579 | |
|
Protein |
rpsA |
PMID:15690043 |
Experiment(s):EBI-887579 | |
|
Protein |
rpsA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
Vsr |
PMID:19376855 |
||
|
Protein |
DnaN: beta sliding clamp of DNA Pol III |
MutL only binds to the beta subunit in the presence of ss DNA. |
PMID:16546997 |
native gel electrophoresis |
|
Protein |
MutH |
data suggest the ATPase fragment of MutL directly contacts MutH |
PMID:9827806 |
Chemical crosslinking |
|
Protein |
MutS |
PMID:9250691 |
Visualized using EM | |
|
Protein |
UvrD |
PMID:9880500 |
2-Hybrid Screen | |
|
Protein |
MutH |
The C-terminal 218 amino acids of MutL were sufficient for the two-hybrid interaction with MutH |
PMID:9880500 |
2 Hybrid Screen |
|
Protein |
UvrD |
Both the N- and C-termini of UvrD were required for interaction with MutL |
PMID:9482750 |
2 Hybrid Screen |
|
Protein |
The N-terminal fragment of MutL (residues 1-349) was sufficient to bind RecA. MutL inhibits the ssDNA-dependent ATPase activity of RecA (Fig. 4) |
PMID:22001225 |
Far-Western and pull-down assays. | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MPIQVLPPQL ANQIAAGEVV ERPASVVKEL VENSLDAGAT RIDIDIERGG AKLIRIRDNG CGIKKDELAL ALARHATSKI ASLDDLEAII SLGFRGEALA SISSVSRLTL TSRTAEQQEA WQAYAEGRDM NVTVKPAAHP VGTTLEVLDL FYNTPARRKF LRTEKTEFNH IDEIIRRIAL ARFDVTINLS HNGKIVRQYR AVPEGGQKER RLGAICGTAF LEQALAIEWQ HGDLTLRGWV ADPNHTTPAL AEIQYCYVNG RMMRDRLINH AIRQACEDKL GADQQPAFVL YLEIDPHQVD VNVHPAKHEV RFHQSRLVHD FIYQGVLSVL QQQLETPLPL DDEPQPAPRS IPENRVAAGR NHFAEPAARE PVAPRYTPAP ASGSRPAAPW PNAQPGYQKQ QGEVYRQLLQ TPAPMQKLKA PEPQEPALAA NSQSFGRVLT IVHSDCALLE RDGNISLLSL PVAERWLRQA QLTPGEAPVC AQPLLIPLRL KVSAEEKSAL EKAQSALAEL GIDFQSDAQH VTIRAVPLPL RQQNLQILIP ELIGYLAKQS VFEPGNIAQW IARNLMSEHA QWSMAQAITL LADVERLCPQ LVKTPPGGLL QSVDLHPAIK ALKDE |
| Length |
615 |
| Mol. Wt |
67.924 kDa |
| pI |
6.8 (calculated) |
| Extinction coefficient |
54,890 - 55,765 (calc based on 11 Y, 7 W, and 7 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
|
Purified protein |
MutL R266E[7] |
|
|
Purified protein |
MutL R95F[7] |
|
|
Purified protein |
MutL N302A[7] |
|
|
Purified protein |
MutL R261H[7] |
|
|
Purified protein |
MutL K307A[7] |
|
|
Purified protein |
MutL E29A[7] |
|
|
Purified protein |
MutL R95F/N302A[7] |
|
|
Purified protein |
MutL I90R[7] |
|
|
Purified protein |
MutL I90E[7] |
|
|
Purified protein |
MutL R237E[7] |
|
|
Purified protein |
MutL G238A[7] |
|
|
Purified protein |
MutL K159E[7] |
|
|
Purified protein |
MutL G238D[7] |
|
|
Purified protein |
MutL R177E[7] |
|
|
Purification protocol |
Single-step purification of His6-MutL |
PMID:8747662 |
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0013655 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG11281 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11281 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120001254 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1258 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
Ecoli K-12 |
6.625+/-0.086 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.027070926 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
160 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
77 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
118 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
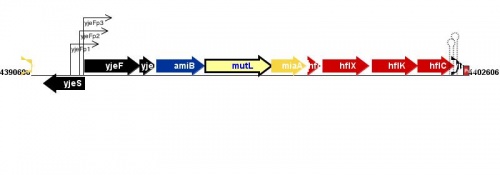 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:4395415..4395455
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for mutL | |
|
microarray |
Summary of data for mutL from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to mutL Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11281 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1258 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11281 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001254 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0013655 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Danio rerio |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Drosophila melanogaster |
|
From Inparanoid:20070104 |
|
Drosophila pseudoobscura |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Macaca mulatta |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Saccharomyces cerevisiae |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Tetraodon nigroviridis |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
MUTL |
From SHIGELLACYC |
|
E. coli O157 |
MUTL |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
PF02518 Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase |
||
|
EcoCyc:EG11281 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11281 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001254 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1258 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0013655 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ 7.00 7.01 7.02 7.03 7.04 7.05 7.06 7.07 7.08 7.09 7.10 7.11 7.12 7.13 Junop, MS et al. (2003) In vitro and in vivo studies of MutS, MutL and MutH mutants: correlation of mismatch repair and DNA recombination. DNA Repair (Amst.) 2 387-405 PubMed
Categories
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Apis mellifera
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Bos taurus
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Canis familiaris
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Danio rerio
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Drosophila melanogaster
- Genes with homologs in Drosophila pseudoobscura
- Genes with homologs in Gallus gallus
- Genes with homologs in Homo sapiens
- Genes with homologs in Macaca mulatta
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Oryza gramene
- Genes with homologs in Pan troglodytes
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Saccharomyces cerevisiae
- Genes with homologs in Schizosaccharomyces pombe
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Tetraodon nigroviridis
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


