mntH:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
mntH |
|---|---|
| Gene Synonym(s) |
ECK2387, b2392, JW2388, mntA, yfeP[1], yfeP |
| Product Desc. |
MntH manganese ion transporter[2][3] Mn(2+) transporter, role in bacterial response to reactive oxygen species; also transports Fe(2+), Cd(2+) and other divalent metals[4] |
| Product Synonyms(s) |
manganese/divalent cation transporter[1], B2392[2][1], MntA[2][1], MntH[2][1], YfeP[2][1] , ECK2387, JW2388, mntA, yfeP, b2392 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Fur and OxyR regulons. Regulated by MntR. NRAMP family.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
mntH |
|---|---|
| Mnemonic |
Mn transport |
| Synonyms |
ECK2387, b2392, JW2388, mntA, yfeP[1], yfeP |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
54.09 minutes, 54.09 minutes |
MG1655: 2510728..2509490 |
||
|
NC_012967: 2446931..2445693 |
||||
|
NC_012759: 2395295..2396533 |
||||
|
W3110 |
|
W3110: 2518152..2516914 |
||
|
DH10B: 2602493..2601255 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔmntH (Keio:JW2388) |
deletion |
deletion |
PMID:16738554 |
||||
|
mntHN37D |
N37D |
Loss of function |
seeded from UniProt:P0A769 | ||||
|
mntHE102D,Q |
E102D,Q |
Loss of function |
seeded from UniProt:P0A769 | ||||
|
mntHG36P |
G36P |
Loss of function |
seeded from UniProt:P0A769 | ||||
|
mntHD109E,N |
D109E,N |
Transports manganese to a very low level |
seeded from UniProt:P0A769 | ||||
|
mntHE112D,Q |
E112D,Q |
Transports manganese to a very low level |
seeded from UniProt:P0A769 | ||||
|
mntHP35N,Q |
P35N,Q |
Loss of function |
seeded from UniProt:P0A769 | ||||
|
mntHD34E,N |
D34E,N |
Loss of function |
seeded from UniProt:P0A769 | ||||
|
mntHG115A,P |
G115A,P |
Loss of function |
seeded from UniProt:P0A769 | ||||
|
mntHE154Q |
E154Q |
Transports manganese to a level half of that of wild-type |
seeded from UniProt:P0A769 | ||||
|
mntHD238N |
D238N |
Transports manganese to a very low level |
seeded from UniProt:P0A769 | ||||
|
ΔmntH729::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW2388 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCACGAACTATCGCGTTGAGAG Primer 2:CCtAATCCCAGCGCCGTCCCCAC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
zfd-1::Tn10 |
Linked marker |
est. P1 cotransduction: 52% [6] | |
|
Linked marker |
est. P1 cotransduction: 98% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G7254 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG14157 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120003828 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB3909 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0007889 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
MntH |
|---|---|
| Synonyms |
manganese/divalent cation transporter[1], B2392[2][1], MntA[2][1], MntH[2][1], YfeP[2][1] , ECK2387, JW2388, mntA, yfeP, b2392 |
| Product description |
MntH manganese ion transporter[2][3] Mn(2+) transporter, role in bacterial response to reactive oxygen species; also transports Fe(2+), Cd(2+) and other divalent metals[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0015292 |
uniporter activity |
PMID:10712688 |
IMP: Inferred from Mutant Phenotype |
F |
complete | |||
|
GO:0015684 |
ferrous iron transport |
PMID:10712688 |
IMP: Inferred from Mutant Phenotype |
P |
preference for Mn(II) |
complete | ||
|
GO:0030001 |
metal ion transport |
PMID:10712688 |
IMP: Inferred from Mutant Phenotype |
P |
Mn(II) and Fe (II) |
complete | ||
|
GO:0005215 |
transporter activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001046 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005886 |
plasma membrane |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00221 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0997 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-1003 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006828 |
manganese ion transport |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00221 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0015293 |
symporter activity |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00221 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0015293 |
symporter activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0769 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005886 |
plasma membrane |
PMID:14607838 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0016021 |
integral to membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0812 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
PMID:14607838 |
IMP: Inferred from Mutant Phenotype |
C |
complete | |||
|
GO:0016021 |
integral to membrane |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-9909 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030001 |
metal ion transport |
PMID:14607838 |
IMP: Inferred from Mutant Phenotype |
P |
Cd(II) |
complete | ||
|
GO:0030001 |
metal ion transport |
PMID:10712688 |
IMP: Inferred from Mutant Phenotype |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0030001 |
metal ion transport |
PMID:14607838 |
IMP: Inferred from Mutant Phenotype |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0015293 |
symporter activity |
PMID:18227061 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0030145 |
manganese ion binding |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00221 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006824 |
cobalt ion transport |
PMID:15641799 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0030145 |
manganese ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0464 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0015691 |
cadmium ion transport |
PMID:18227061 |
IDA: Inferred from Direct Assay |
P |
complete | |||
|
GO:0006826 |
iron ion transport |
PMID:15641799 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0006828 |
manganese ion transport |
PMID:15641799 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0015691 |
cadmium ion transport |
PMID:15641799 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0015078 |
hydrogen ion transmembrane transporter activity |
PMID:15641799 |
IMP: Inferred from Mutant Phenotype |
F |
complete | |||
|
GO:0005384 |
manganese ion transmembrane transporter activity |
PMID:15641799 |
IMP: Inferred from Mutant Phenotype |
F |
complete | |||
|
GO:0015086 |
cadmium ion transmembrane transporter activity |
PMID:15641799 |
IMP: Inferred from Mutant Phenotype |
F |
complete | |||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
plasma membrane |
C-terminus localized to the periplasm with 11 predicted transmembrane domains |
Daley et al. (2005) [7] |
||
|
plasma membrane |
From EcoCyc[3] |
| ||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MTNYRVESSS GRAARKMRLA LMGPAFIAAI GYIDPGNFAT NIQAGASFGY QLLWVVVWAN LMAMLIQILS AKLGIATGKN LAEQIRDHYP RPVVWFYWVQ AEIIAMATDL AEFIGAAIGF KLILGVSLLQ GAVLTGIATF LILMLQRRGQ KPLEKVIGGL LLFVAAAYIV ELIFSQPNLA QLGKGMVIPS LPTSEAVFLA AGVLGATIMP HVIYLHSSLT QHLHGGSRQQ RYSATKWDVA IAMTIAGFVN LAMMATAAAA FHFSGHTGVA DLDEAYLTLQ PLLSHAAATV FGLSLVAAGL SSTVVGTLAG QVVMQGFIRF HIPLWVRRTV TMLPSFIVIL MGLDPTRILV MSQVLLSFGI ALALVPLLIF TSDSKLMGDL VNSKRVKQTG WVIVVLVVAL NIWLLVGTAL GL |
| Length |
412 |
| Mol. Wt |
44.194 kDa |
| pI |
10.2 (calculated) |
| Extinction coefficient |
57,410 (calc based on 9 Y, 8 W, and 0 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0007889 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:G7254 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG14157 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120003828 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB3909 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
136 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
58 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
57 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
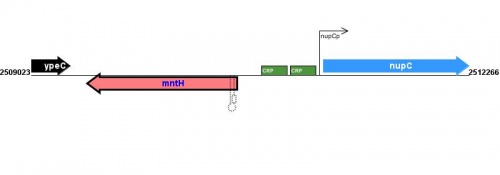 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:2510708..2510748
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for mntH | |
|
microarray |
Summary of data for mntH from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to mntH Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G7254 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB3909 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG14157 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120003828 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0007889 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Danio rerio |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Drosophila melanogaster |
|
From Inparanoid:20070104 |
|
Drosophila pseudoobscura |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Macaca mulatta |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Saccharomyces cerevisiae |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Tetraodon nigroviridis |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
MNTH |
From SHIGELLACYC |
|
E. coli O157 |
Z3658 |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G7254 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG14157 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120003828 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB3909 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0007889 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Daley, DO et al. (2005) Global topology analysis of the Escherichia coli inner membrane proteome. Science 308 1321-3 PubMed
Categories
- TMHMM Prediction
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Apis mellifera
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Bos taurus
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Canis familiaris
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Danio rerio
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Drosophila melanogaster
- Genes with homologs in Drosophila pseudoobscura
- Genes with homologs in Gallus gallus
- Genes with homologs in Homo sapiens
- Genes with homologs in Macaca mulatta
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Oryza gramene
- Genes with homologs in Pan troglodytes
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Saccharomyces cerevisiae
- Genes with homologs in Schizosaccharomyces pombe
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Tetraodon nigroviridis
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157



