hscA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
hscA |
|---|---|
| Gene Synonym(s) | |
| Product Desc. |
chaperone, member of Hsp70 protein family[2][3] Hsc66, DnaK-like chaperone, specific for IscU; involved in FtsZ-ring formation; HscB is the J-like co-chaperone for HscA[4] |
| Product Synonyms(s) |
DnaK-like molecular chaperone specific for IscU[1], B2526[2][1], Hsc[2][1], HscA[2][1], Hsc66[2][1] , ECK2523, hsc, JW2510, b2526 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
dnaK, hscA, hscC and yegD are paralogs.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
hscA |
|---|---|
| Mnemonic |
Heat shock cognate |
| Synonyms | |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
57.23 minutes |
MG1655: 2656957..2655107 |
||
|
NC_012967: 2579634..2577784 |
||||
|
NC_012759: 2540912..2542762 |
||||
|
W3110 |
|
W3110: 2657591..2655741 |
||
|
DH10B: 2748722..2746872 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔhscA (Keio:JW2510) |
deletion |
deletion |
PMID:16738554 |
||||
|
hscAS216F |
S216F |
(in hsca1) |
Strain variation; seeded from UniProt:P0A6Z1 | ||||
|
ΔhscA772::kan |
PMID:16738554 |
||||||
|
hscA |
deletion |
Sensitivity to |
increases sensitivity to bicyclomycin |
PMID:21357484 |
fig 2 | ||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW2510 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCGCCTTATTACAAATTAGTGA Primer 2:CCAACCTCGTCCACGGAATGGC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 4% [6] | ||
|
Linked marker |
est. P1 cotransduction: 76% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG12130 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12130 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120002032 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB2051 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0008315 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
HscA |
|---|---|
| Synonyms |
DnaK-like molecular chaperone specific for IscU[1], B2526[2][1], Hsc[2][1], HscA[2][1], Hsc66[2][1] , ECK2523, hsc, JW2510, b2526 |
| Product description |
chaperone, member of Hsp70 protein family[2][3] Hsc66, DnaK-like chaperone, specific for IscU; involved in FtsZ-ring formation; HscB is the J-like co-chaperone for HscA[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0005515 |
protein binding |
PMID:11983079 |
IPI: Inferred from Physical Interaction |
UniProtKB:P0ACD4 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005515 |
protein binding |
PMID:12871959 |
IPI: Inferred from Physical Interaction |
UniProtKB:P0ACD4 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005515 |
protein binding |
PMID:15485839 |
IPI: Inferred from Physical Interaction |
UniProtKB:P0A6L9 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005515 |
protein binding |
PMID:19821612 |
IPI: Inferred from Physical Interaction |
EcoCyc:BIOTIN-SYN-CPLX |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001023 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR010236 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
PMID:10713091 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0005524 |
ATP binding |
PMID:11323718 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0006457 |
protein folding |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00679 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006457 |
protein folding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR010236 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016226 |
iron-sulfur cluster assembly |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR010236 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016226 |
iron-sulfur cluster assembly |
PMID:10393315 |
IGI: Inferred from Genetic Interaction |
P |
Seeded from EcoCyc (v14.0) |
Missing: with/from | ||
|
GO:0016226 |
iron-sulfur cluster assembly |
PMID:10544286 |
IMP: Inferred from Mutant Phenotype |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0016887 |
ATPase activity |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00679 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016887 |
ATPase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR010236 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016887 |
ATPase activity |
PMID:10713091 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0043531 |
ADP binding |
PMID:11323718 |
IDA: Inferred from Direct Assay |
F |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0051082 |
unfolded protein binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR010236 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0070417 |
cellular response to cold |
PMID:7665466 |
IEP: Inferred from Expression Pattern |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
rplJ |
PMID:15690043 |
Experiment(s):EBI-878924, EBI-882104 | |
|
Protein |
secA |
PMID:15690043 |
Experiment(s):EBI-878924 | |
|
Protein |
ygeQ |
PMID:15690043 |
Experiment(s):EBI-882104 | |
|
Protein |
pepB |
PMID:15690043 |
Experiment(s):EBI-882104 | |
|
Protein |
rplL |
PMID:15690043 |
Experiment(s):EBI-882104 | |
|
Protein |
rplT |
PMID:15690043 |
Experiment(s):EBI-882104 | |
|
Protein |
ugpB |
PMID:15690043 |
Experiment(s):EBI-882104 | |
|
Protein |
rplQ |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
rpsE |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
ahpC |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
rplE |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
cheZ |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
rpsC |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
cedA |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
chbA |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
rpsI |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
rpsD |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
cybC |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
ygjQ |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
rplB |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
rplF |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
gnsA |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
rplN |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
fliT |
PMID:16606699 |
Experiment(s):EBI-1143130 | |
|
Protein |
rplJ |
PMID:19402753 |
LCMS(ID Probability):99.0 MALDI(Z-score):17.053160 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MALLQISEPG LSAAPHQRRL AAGIDLGTTN SLVATVRSGQ AETLADHEGR HLLPSVVHYQ QQGHSVGYDA RTNAALDTAN TISSVKRLMG RSLADIQQRY PHLPYQFQAS ENGLPMIETA AGLLNPVRVS ADILKALAAR ATEALAGELD GVVITVPAYF DDAQRQGTKD AARLAGLHVL RLLNEPTAAA IAYGLDSGQE GVIAVYDLGG GTFDISILRL SRGVFEVLAT GGDSALGGDD FDHLLADYIR EQAGIPDRSD NRVQRELLDA AIAAKIALSD ADSVTVNVAG WQGEISREQF NELIAPLVKR TLLACRRALK DAGVEADEVL EVVMVGGSTR VPLVRERVGE FFGRPPLTSI DPDKVVAIGA AIQADILVGN KPDSEMLLLD VIPLSLGLET MGGLVEKVIP RNTTIPVARA QDFTTFKDGQ TAMSIHVMQG ERELVQDCRS LARFALRGIP ALPAGGAHIR VTFQVDADGL LSVTAMEKST GVEASIQVKP SYGLTDSEIA SMIKDSMSYA EQDVKARMLA EQKVEAARVL ESLHGALAAD AALLSAAERQ VIDDAAAHLS EVAQGDDVDA IEQAIKNVDK QTQDFAARRM DQSVRRALKG HSVDEV |
| Length |
616 |
| Mol. Wt |
65.652 kDa |
| pI |
4.9 (calculated) |
| Extinction coefficient |
20,400 - 20,650 (calc based on 10 Y, 1 W, and 2 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0008315 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG12130 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12130 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120002032 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2051 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
4.91E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
E. coli K-12 MG1655 |
2256 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
542 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
2252 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
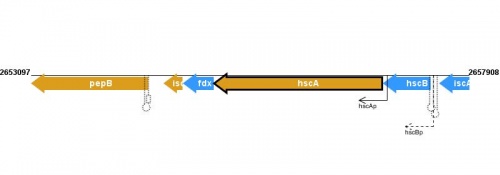 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:2656937..2656977
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for hscA | |
|
microarray |
Summary of data for hscA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to hscA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG12130 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2051 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12130 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120002032 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0008315 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
HSCA |
From SHIGELLACYC |
|
E. coli O157 |
HSCA |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG12130 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12130 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120002032 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2051 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0008315 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories


