gyrA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
gyrA |
|---|---|
| Gene Synonym(s) |
ECK2223, b2231, JW2225, hisW, nalA, parD, nfxA, norA[1], norA |
| Product Desc. |
Component of DNA gyrase[3] DNA gyrase, subunit A; nalidixic acid resistance; cold shock regulon[4] |
| Product Synonyms(s) |
DNA gyrase (type II topoisomerase), subunit A[1], B2231[2][1], ParD[2][1], NorA[2][1], NfxA[2][1], NalA[2][1], HisW[2][1], GyrA[2][1] , ECK2223, hisW, JW2225, nalA, nfxA, norA, parD, b2231 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
gyrA is an essential gene. HT_Cmplx22_Cyt: DnaK+GrpE+GyrA+RpsA.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
gyrA |
|---|---|
| Mnemonic |
Gyrase |
| Synonyms |
ECK2223, b2231, JW2225, hisW, nalA, parD, nfxA, norA[1], norA |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
50.32 minutes |
MG1655: 2337442..2334815 |
||
|
NC_012967: 2286120..2283493 |
||||
|
NC_012759: 2220620..2223247 |
||||
|
W3110 |
|
W3110: 2344090..2341463 |
||
|
DH10B: 2428430..2425803 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
|
Coding Start (SO:0000323) |
2334818 |
Edman degradation |
PMID:3029031 |
| |
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
gyrAA67S |
A67S |
Resistant to |
(in PPA-10; quinolone-resistant) |
Strain variation; seeded from UniProt:P0AES4 | |||
|
gyrAG81C |
G81C |
(in NAL-97; quinolone-resistant) |
Strain variation; seeded from UniProt:P0AES4 | ||||
|
gyrAD678E |
D678E |
(in strain: 227) |
Strain variation; seeded from UniProt:P0AES4 | ||||
|
gyrAI798IMMI |
I798IMMI |
(in KL-16; quinolone- resistant) |
Strain variation; seeded from UniProt:P0AES4 | ||||
|
gyrAA828S |
A828S |
(in strain: 227) |
Strain variation; seeded from UniProt:P0AES4 | ||||
|
gyrAS83A |
S83A |
Resistant to fluoroquinolones |
seeded from UniProt:P0AES4 | ||||
|
gyrAQ106R |
Q106R |
Resistant to fluoroquinolones |
seeded from UniProt:P0AES4 | ||||
|
gyrAS83W |
S83W |
(in PPA-18 and strain 227; quinolone-resistant) |
Strain variation; seeded from UniProt:P0AES4 | ||||
|
gyrAA84P |
A84P |
(in PPA-05; quinolone-resistant) |
Strain variation; seeded from UniProt:P0AES4 | ||||
|
gyrAD87N |
GAC -> AAC at codon 87 |
D87N |
Resistant to |
(in NAL-113 and KL-16; quinolone- resistant) resistant to 200 ug/ml nalidixic acid |
Strain variation; seeded from UniProt:P0AES4 | ||
|
gyrAQ106H |
Q106H |
(in NAL-89; quinolone-resistant) |
Strain variation; seeded from UniProt:P0AES4 | ||||
|
gyrAD87V |
D87V |
(in strain: 202; quinolone- resistant) |
Strain variation; seeded from UniProt:P0AES4 | ||||
|
gyrAS83L |
S83L |
(in NAL-51, NAL-112, NAL-118 and NAL-119; quinolone-resistant) |
Strain variation; seeded from UniProt:P0AES4 | ||||
|
gyrA98(NalR) |
nalidixic acid resistant |
||||||
|
gyrA20(NalR) |
nalidixic acid resistant |
||||||
|
gyrA12(NalR) |
nalidixic acid resistant |
||||||
|
gyrA26(NalR) |
nalidixic acid resistant |
||||||
|
gyrA39(NalR) |
nalidixic acid resistant |
||||||
|
gyrA96(NalR) |
nalidixic acid resistant |
||||||
|
gyrA91(NalR) |
nalidixic acid resistant |
||||||
|
gyrA93(NalR) |
nalidixic acid resistant |
||||||
|
gyrA216(NalR) |
nalidixic acid resistant |
||||||
|
gyrA220(NalR) |
nalidixic acid resistant |
||||||
|
gyrA218(NalR) |
nalidixic acid resistant |
||||||
|
gyrA13(NalR) |
nalidixic acid resistant |
||||||
|
gyrA19(NalR) |
nalidixic acid resistant |
||||||
|
gyrA111(NalR) |
nalidixic acid resistant |
||||||
|
gyrA261(NalR) |
nalidixic acid resistant |
||||||
|
gyrA90(NalR) |
nalidixic acid resistant |
||||||
|
gyrA99(NalR) |
nalidixic acid resistant |
||||||
|
gyrA49 |
|||||||
|
gyrA217(NalR) |
nalidixic acid resistant |
||||||
|
gyrA223(NalR) |
nalidixic acid resistant |
||||||
|
gyrA219(NalR) |
nalidixic acid resistant |
||||||
|
gyrA271(NalR) |
nalidixic acid resistant |
||||||
|
gyrA260(NalR) |
nalidixic acid resistant |
||||||
|
gyrA270(NalR) |
nalidixic acid resistant |
||||||
|
[gyrA43](ts) |
temperature sensitive |
||||||
|
gyrA25(NalR) |
nalidixic acid resistant |
||||||
|
gyrA21(NalR) |
nalidixic acid resistant |
||||||
|
gyrA222(NalR) |
nalidixic acid resistant |
||||||
|
gyrA29(NalR) |
nalidixic acid resistant |
||||||
|
gyrA17(NalR) |
nalidixic acid resistant |
||||||
|
gyrA42(ts) |
temperature sensitive |
||||||
|
gyrA283(NalR) |
nalidixic acid resistant |
||||||
|
gyrA44(ts) |
temperature sensitive |
||||||
|
gyrA37(NalR) |
nalidixic acid resistant |
||||||
|
gyrA329(NalR) |
nalidixic acid resistant |
||||||
|
gyrA284(NalR) |
nalidixic acid resistant |
||||||
|
gyrA900 |
|||||||
|
gyrA586(NalR) |
nalidixic acid resistant |
||||||
|
gyrA(Am) from OV6 |
Growth Phenotype |
at 42C the separation of DNA into nucleoids and division localization are both disturbed but cells have continued growth and DNA replication production of cells, and normal sized cells are all produced at 42C |
PMID:330757 |
||||
|
gyrA43(TS) |
Resistant to |
Resistant to Nalidixic Acid at the permissive temperature 32 C. |
PMID:3025879 |
Strain: KRF949 Parental Strain: CGSC KNK43
|
|||
|
gyrA43 |
Resistant to |
Resistant to Triazolealanine. |
PMID:3025879 |
Strain: KRE449 Parental:CGSC KNK453 |
Results on table 4. | ||
|
nalA mutation in MH5 |
Resistant to |
resistance to nalidixic acid |
PMID:4895844 |
similar strain: MH7 resistant at 40ug nal per ml, sensitive at 60ug nal per ml | |||
|
gyrAR S83L |
S83L |
Resistant to |
Quinolone Resistance |
PMID:8524852 |
The mutation conferred an increase in resistance toward Quinolones. See figure 1B. chEBI:23765 | ||
|
gyrAnal-112 |
TCG-->TTG |
S83L |
Resistant to |
Resistant to nalidixic acid |
PMID:2168148 |
Parental strain: KL16 |
Results found in Table 1. Mutation and phenotype identical in strains gyrAnal-118, gyrAnal-119, and gyrAnal-51. |
|
gyrAppa-18 |
TCG-->TGG |
S83W |
Resistant to |
Resistant to pipemidic acid. |
PMID:2168148 |
Parental strain: KL16 |
Results found in Table 1. |
|
gyrAnal-113 |
GAC-->AAC |
D87N |
Resistant to |
Resistant to nalidixic acid |
PMID:2168148 |
Parental strain: KL16 |
Results found in Table 1 |
|
gyrAnal-97 |
GGT-->TGT |
G81C |
Resistant to |
Resistant to nalidixic acid. |
PMID:2168148 |
Parental strain: KL16 |
Results found in Table 1. |
|
gyrAppa-5 |
GCG-->CCG |
A84P |
Resistant to |
Resistant to pipemidic acid. |
PMID 2168148 |
Parental strain: KL16 |
Results found in Table 1. |
|
gyrAppa-10 |
GGC-->TCC |
A67S |
Resistant to |
Resistant to pipemidic acid. |
PMID:2168148 |
Parental strain: KL16 |
Results found in Table 1. |
|
gyrAnal-89 |
CAG-->CAT |
Q106H |
Resistant to |
Resistant to nalidixic acid. |
PMID:2168148 |
Parental strain: KL16 |
Results found in Table 1. |
| edit table |
<protect></protect>
Notes
A Latin American study found that the gyrA single-point mutation has caused E. coli to become resistant to Fluoroquinolones, or antimicrobial drugs. This has resulted in an increase in the number of urinary tract infections. [5]
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW2225 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAGCGACCTTGCGAGAGAAAT Primer 2:CCTTCTTCTTCTGGCTCGTCGTC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 59% [7] | ||
|
Linked marker |
est. P1 cotransduction: 76% [7] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10423 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10423 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000416 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0418 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0007370 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
GyrA |
|---|---|
| Synonyms |
DNA gyrase (type II topoisomerase), subunit A[1], B2231[2][1], ParD[2][1], NorA[2][1], NfxA[2][1], NalA[2][1], HisW[2][1], GyrA[2][1] , ECK2223, hisW, JW2225, nalA, nfxA, norA, parD, b2231 |
| Product description |
Component of DNA gyrase[3] DNA gyrase, subunit A; nalidixic acid resistance; cold shock regulon[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0042493 |
response to drug |
PMID:6178722 |
IMP: Inferred from Mutant Phenotype |
P |
complete | |||
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003918 |
DNA topoisomerase (ATP-hydrolyzing) activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002205 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003918 |
DNA topoisomerase (ATP-hydrolyzing) activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005743 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003918 |
DNA topoisomerase (ATP-hydrolyzing) activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013757 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003918 |
DNA topoisomerase (ATP-hydrolyzing) activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013758 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003918 |
DNA topoisomerase (ATP-hydrolyzing) activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013760 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003918 |
DNA topoisomerase (ATP-hydrolyzing) activity |
PMID:368801 |
IDA: Inferred from Direct Assay |
F |
With GyrB Purified E. coli gyrase was shown to induce cleavage and supercoiling in ColE1 DNA in the presence of ATP, but not after the addition of novobiocin or coumermycin A1 (Figure 2). Supercoiling of DNA was monitored by agarose gel electrophoresis (Figure 2A), while the hydrolysis of ATP was measured by following the release of 32-Pi measured by thin layer chromatography (Figure 2B). |
complete | ||
|
GO:0003677 |
DNA binding |
PMID:6294616 |
IDA: Inferred from Direct Assay |
F |
E. coli gyrase incubated with radiolabeled ColE1 DNA and passed over nitrocellulose filters. The complexes were retained on the filters and radioactivity of the filters was measured and reported in Figure 1. |
complete | ||
|
GO:0003918 |
DNA topoisomerase (ATP-hydrolyzing) activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:5.99.1.3 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0042493 |
response to drug |
PMID:770163 |
IMP: Inferred from Mutant Phenotype |
P |
Oxolinic acid (Quinolone) |
complete | ||
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002205 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005743 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0042493 |
response to drug |
PMID:4208771 |
IMP: Inferred from Mutant Phenotype |
P |
Naladixic Acid (Quinolone) |
complete | ||
|
GO:0003677 |
DNA binding |
PMID:1851291 |
IDA: Inferred from Direct Assay |
F |
The C-terminal domain of GyrA (572-857aa) was shown to bind and retard the movement of a 147bp fragment of ColE1 DNA through a 5% polyacrylamide gel (Figure 2). |
complete | ||
|
GO:0006351 |
transcription, DNA-dependent |
PMID:347446 |
IDA: Inferred from Direct Assay |
P |
E. coli gyrase consists of the gene products of gyrA and gyrB genes (Figures 2 & 7). |
complete | ||
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006691 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008094 |
DNA-dependent ATPase activity |
PMID:6094559 |
IDA: Inferred from Direct Assay |
F |
GyrA and DNA (bound to GyrA) are required for ATPase activity of GyrB (Figure 1). |
complete | ||
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013757 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006265 |
DNA topological change |
PMID:6327603 |
IMP: Inferred from Mutant Phenotype |
P |
Nucleoids from E. coli cells containing DNA labeled with [methyl-3H]thymidine were isolated and separated using an ethidium bromide-containing sucrose density gradient. The sedimentation profiles were reported for gyrA and gyrB temperature-sensitive mutants at the permissive and non-permissive temperatures (Figure 1 & 2). The ethidium bromide concentration is proportional to superhelical density, meaning that less supercoiled DNA from a gyrase mutant grown at the non-permissive temperature requires a lower concentration of ethidium bromide. |
complete | ||
|
GO:0006351 |
transcription, DNA-dependent |
PMID:15535863 |
IMP: Inferred from Mutant Phenotype |
P |
The authors used 3 conditions (treatment with either a quinolone or coumarin anitbiotic or growth of a gyrAts mutant at non-permissive temperature) to alter negative supercoiling of DNA and measured the changes in gene expression by microarray analysis. More than 300 SSGs (supercoiling sensitive genes) were identified that responded rapidly to the experimental conditions. |
complete | ||
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013758 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
PMID:7952188 |
IDA: Inferred from Direct Assay |
C |
Immunogold labeling of cells with anti-GyrA antibodies indicated the presence of GyrA in the cytoplasm of E. coli. |
complete | ||
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013760 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005694 |
chromosome |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002205 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005694 |
chromosome |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005743 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005694 |
chromosome |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006691 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005694 |
chromosome |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013757 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005694 |
chromosome |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013758 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005694 |
chromosome |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013760 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
PMID:7952188 |
IDA: Inferred from Direct Assay |
C |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0006261 |
DNA-dependent DNA replication |
PMID:227840 |
IMP: Inferred from Mutant Phenotype |
P |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0006265 |
DNA topological change |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002205 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006265 |
DNA topological change |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005743 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006265 |
DNA topological change |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006691 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006265 |
DNA topological change |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013757 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006265 |
DNA topological change |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013758 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006265 |
DNA topological change |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR013760 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009330 |
DNA topoisomerase complex (ATP-hydrolyzing) |
PMID:347446 |
IDA: Inferred from Direct Assay |
C |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0016020 |
membrane |
PMID:16858726 |
IDA: Inferred from Direct Assay |
C |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0046677 |
response to antibiotic |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0046 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of DNA gyrase |
could be indirect |
||
|
Protein |
ycgC |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
aceF |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
add |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
clpA |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
dnaJ |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
dnaK |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
ffh |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
ftsZ |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
glyS |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
zraS |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
lpdA |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
malK |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
metK |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
pstB |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
recA |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
rho |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
rpsB |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
tufA |
PMID:15690043 |
Experiment(s):EBI-884206, EBI-891237 | |
|
Protein |
yjfZ |
PMID:15690043 |
Experiment(s):EBI-884206 | |
|
Protein |
argS |
PMID:16606699 |
Experiment(s):EBI-1142253 | |
|
Protein |
idnO |
PMID:16606699 |
Experiment(s):EBI-1142253 | |
|
Protein |
rpsB |
PMID:16606699 |
Experiment(s):EBI-1142253 | |
|
Protein |
yfaS |
PMID:16606699 |
Experiment(s):EBI-1142253 | |
|
Protein |
aspC |
PMID:16606699 |
Experiment(s):EBI-1142253 | |
|
Protein |
nrdI |
PMID:16606699 |
Experiment(s):EBI-1142253 | |
|
Protein |
rtcB |
PMID:16606699 |
Experiment(s):EBI-1142253 | |
|
Protein |
rho |
PMID:19402753 |
MALDI(Z-score):18.636092 | |
|
Protein |
clpA |
PMID:19402753 |
MALDI(Z-score):19.599720 | |
|
Protein |
pstB |
PMID:19402753 |
MALDI(Z-score):35.274106 | |
|
Protein |
recA |
PMID:19402753 |
MALDI(Z-score):29.421963 | |
|
Protein |
metK |
PMID:19402753 |
MALDI(Z-score):37.360726 | |
|
Protein |
add |
PMID:19402753 |
MALDI(Z-score):32.777259 | |
|
Protein |
dnaJ |
PMID:19402753 |
MALDI(Z-score):34.861527 | |
|
Protein |
tufB |
PMID:19402753 |
MALDI(Z-score):22.155359 | |
|
Protein |
mreB |
PMID:19402753 |
MALDI(Z-score):36.098785 | |
|
Protein |
The binding of gyrase to DNA: analysis by retention by nitrocellulose filters. Higgins & Cozzarelli (1982)[8] |
|||
|
Small Molecule |
Naladixic Acid |
Studies on the mechanism of action of nalidixic acid. Bourguignon et al. (1973) [10] |
||
|
Small Molecule |
Oxolinic acid |
Replication of Escherichia coli DNA in vitro: inhibition by oxolinic acid. Staudenbauer (1976) [11] |
||
|
DNA |
DNA |
The C-terminal domain of GyrA is sufficient to bind DNA (residues 572-875). |
The C-terminal domain of the Escherichia coli DNA gyrase A subunit is a DNA-binding protein. Reece & Maxwell (1991) [12] |
|
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MSDLAREITP VNIEEELKSS YLDYAMSVIV GRALPDVRDG LKPVHRRVLY AMNVLGNDWN KAYKKSARVV GDVIGKYHPH GDSAVYDTIV RMAQPFSLRY MLVDGQGNFG SIDGDSAAAM RYTEIRLAKI AHELMADLEK ETVDFVDNYD GTEKIPDVMP TKIPNLLVNG SSGIAVGMAT NIPPHNLTEV INGCLAYIDD EDISIEGLME HIPGPDFPTA AIINGRRGIE EAYRTGRGKV YIRARAEVEV DAKTGRETII VHEIPYQVNK ARLIEKIAEL VKEKRVEGIS ALRDESDKDG MRIVIEVKRD AVGEVVLNNL YSQTQLQVSF GINMVALHHG QPKIMNLKDI IAAFVRHRRE VVTRRTIFEL RKARDRAHIL EALAVALANI DPIIELIRHA PTPAEAKTAL VANPWQLGNV AAMLERAGDD AARPEWLEPE FGVRDGLYYL TEQQAQAILD LRLQKLTGLE HEKLLDEYKE LLDQIAELLR ILGSADRLME VIREELELVR EQFGDKRRTE ITANSADINL EDLITQEDVV VTLSHQGYVK YQPLSEYEAQ RRGGKGKSAA RIKEEDFIDR LLVANTHDHI LCFSSRGRVY SMKVYQLPEA TRGARGRPIV NLLPLEQDER ITAILPVTEF EEGVKVFMAT ANGTVKKTVL TEFNRLRTAG KVAIKLVDGD ELIGVDLTSG EDEVMLFSAE GKVVRFKESS VRAMGCNTTG VRGIRLGEGD KVVSLIVPRG DGAILTATQN GYGKRTAVAE YPTKSRATKG VISIKVTERN GLVVGAVQVD DCDQIMMITD AGTLVRTRVS EISIVGRNTQ GVILIRTAED ENVVGLQRVA EPVDEEDLDT IDGSAAEGDD EIAPEVDVDD EPEEE |
| Length |
875 |
| Mol. Wt |
96.964 kDa |
| pI |
5.0 (calculated) |
| Extinction coefficient |
52,260 - 52,760 (calc based on 24 Y, 3 W, and 4 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0007370 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10423 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10423 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000416 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0418 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
1.21E+03 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
68.835+/-0.445 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.241910925 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
5219 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
1407 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
2680 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
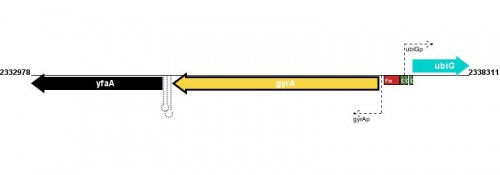 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:2337422..2337462
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for gyrA | |
|
microarray |
Summary of data for gyrA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Accessions Related to gyrA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10423 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0418 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10423 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000416 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0007370 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Macaca mulatta |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
GYRA |
From SHIGELLACYC |
|
E. coli O157 |
GYRA |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10423 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10423 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000416 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0418 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0007370 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ Gales, AC et al. (2000) Occurrence of single-point gyrA mutations among ciprofloxacin-susceptible Escherichia coli isolates causing urinary tract infections in Latin America. Diagn. Microbiol. Infect. Dis. 36 61-4 PubMed
- ↑ 6.0 6.1 CGSC: The Coli Genetics Stock Center
- ↑ 7.0 7.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Higgins, NP & Cozzarelli, NR (1982) The binding of gyrase to DNA: analysis by retention by nitrocellulose filters. Nucleic Acids Res. 10 6833-47 PubMed
- ↑ Higgins, NP et al. (1978) Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc. Natl. Acad. Sci. U.S.A. 75 1773-7 PubMed
- ↑ Bourguignon, GJ et al. (1973) Studies on the mechanism of action of nalidixic acid. Antimicrob. Agents Chemother. 4 479-86 PubMed
- ↑ Staudenbauer, WL (1976) Replication of Escherichia coli DNA in vitro: inhibition by oxolinic acid. Eur. J. Biochem. 62 491-7 PubMed
- ↑ Reece, RJ & Maxwell, A (1991) The C-terminal domain of the Escherichia coli DNA gyrase A subunit is a DNA-binding protein. Nucleic Acids Res. 19 1399-405 PubMed
Categories
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Bos taurus
- Genes with homologs in Macaca mulatta
- Genes with homologs in Oryza gramene
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


