ypjA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
ypjA |
|---|---|
| Gene Synonym(s) |
ECK2644, b2647, JW5422[1], JW5422 |
| Product Desc. |
adhesin-like autotransporter[2][3] Overexpression increases adhesion, function unknown; OM autotransporter homolog[4] |
| Product Synonyms(s) |
adhesin-like autotransporter[1], B2647[2][1], YpjA[2][1] , ECK2644, JW5422, b2647 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Mutants have reduced OM vesiculation (McBroom, 2006).[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
ypjA |
|---|---|
| Mnemonic |
Systematic nomenclature |
| Synonyms |
ECK2644, b2647, JW5422[1], JW5422 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
59.84 minutes |
MG1655: 2780748..2776168 |
||
|
NC_012759: 2661980..2666560 |
||||
|
W3110 |
|
W3110: 2781382..2776802 |
||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔypjA (Keio:JW5422) |
deletion |
deletion |
PMID:16738554 |
||||
|
ΔypjA732::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW5422 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAACAGGACCAGTCCCTATTA Primer 2:CCGAACGACCAGTTCACACCAGC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 20% [6] | ||
|
Linked marker |
est. P1 cotransduction: 10% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G7382 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG13213 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120003950 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB3005 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0008713 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
YpjA |
|---|---|
| Synonyms |
adhesin-like autotransporter[1], B2647[2][1], YpjA[2][1] , ECK2644, JW5422, b2647 |
| Product description |
adhesin-like autotransporter[2][3] Overexpression increases adhesion, function unknown; OM autotransporter homolog[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-1003 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007155 |
cell adhesion |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003991 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009279 |
cell outer membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0998 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0472 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0019867 |
outer membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006315 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0019898 |
extrinsic to membrane |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-9903 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MNTIHLRCLF RMNPLVWCLW ADVAAKLRSL KRYSVFTFQR MKFMNRTSPY YCRRSVLSLL ISALIYAPPG MAAFTTNVIG VVNDETVDGN QKVDERGTTN NTHIINHGQQ NVHGGVSNGS LIESGGYQDI GSHNNFVGQA NNTTINGGRQ SIHDGGISTG TTIESGNQDV YKGGISNGTT IKGGASRVEG GSANGILIDG GSQIVKVQGH ADGTTINKSG SQDVVQGSLA TNTTINGGRQ YVEQSTVETT TIKNGGEQRV YESRALDTTI EGGTQSLNSK STAKNTHIYS GGTQIVDNTS TSDVIEVYSG GVLDVRGGTA TNVTQHDGAI LKTNTNGTTV SGTNSEGAFS IHNHVADNVL LENGGHLDIN AYGSANKTII KDKGTMSVLT NAKADATRID NGGVMDVAGN ATNTIINGGT QNINNYGIAT GTNINSGTQN IKSGGKADTT IISSGSRQVV EKDGTAIGSN ISAGGSLIVY TGGIAHGVNQ ETGSALVANT GAGTDIEGYN KLSHFTITGG EANYVVLENT GELTVVAKTS AKNTTIDTGG KLIVQKEAKT DSTRLNNGGV LEVQDGGEAK HVEQQSGGAL IASTTSGTLI EGTNSYGDAF YIRNSEAKNV VLENAGSLTV VTGSRAVDTI INANGKMDVY GKDVGTVLNS AGTQTIYASA TSDKANIKGG KQTVYGLATE ANIESGEQIV DGGSTEKTHI NGGTQTVQNY GKAINTDIVS GLQQIMANGT AEGSIINGGS QVVNEGGLAE NSVLNDGGTL DVREKGSATG IQQSSQGALV ATTRATRVTG TRADGVAFSI EQGAANNILL ANGGVLTVES DTSSDKTQVN MGGREIVKTK ATATGTTLTG GEQIVEGVAN ETTINDGGIQ TVSANGEAIK TKINEGGTLT VNDNGKATDI VQNSGAALQT STANGIEISG THQYGTFSIS GNLATNMLLE NGGNLLVLAG TEARDSTVGK GGAMQNLGQD SATKVNSGGQ YTLGRSKDEF QALARAEDLQ VAGGTAIVYA GTLADASVSG ATGSLSLMTP RDNVTPVKLE GAVRITDSAT LTLGNGVDTT LADLTAASRG SVWLNSNNSC AGTSNCEYRV NSLLLNDGDV YLSAQTAAPA TTNGIYNTLT TNELSGSGNF YLHTNVAGSR GDQLVVNNNA TGNFKIFVQD TGVSPQSDDA MTLVKTGGGD ASFTLGNTGG FVDLGTYEYV LKSDGNSNWN LTNDVKPNPD PIPNPKPDPK PDPKPDPNPK PDPTPDPTPT PVPEKRITPS TAAVLNMAAT LPLVFDAELN SIRERLNIMK ASPHNNNVWG ATYNTRNNVT TDAGAGFEQT LTGMTVGIDS RNDIPEGITT LGAFMGYSHS HIGFDRGGHG SVGSYSLGGY ASWEHESGFY LDGVVKLNRF KSNVAGKMSS GGAANGSYHS NGLGGHIETG MRFTDGNWNL TPYASLTGFT ADNPEYHLSN GMKSKSVDTR SIYRELGATL SYNMRLGNGM EVEPWLKAAV RKEFVDDNRV KVNSDGNFVN YLSGRRGIYQ AGIKASFSST LSGHLGVGYS HSAGVESPWN AVAGVNWSF |
| Length |
1,569 |
| Mol. Wt |
162.773 kDa |
| pI |
5.5 (calculated) |
| Extinction coefficient |
119,070 - 119,695 (calc based on 43 Y, 10 W, and 5 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0008713 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:G7382 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG13213 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120003950 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB3005 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
2 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
4 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
0a |
molecules/cell/generation |
|
Ribosome Profiling |
Low confidence in the sequencing data set. |
PMID: 24766808 |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
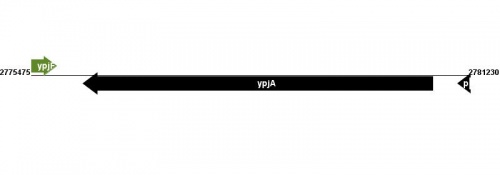 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:2780728..2780768
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for ypjA | |
|
microarray |
Summary of data for ypjA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (2780805..2781122) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to ypjA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G7382 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB3005 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG13213 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120003950 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0008713 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
E. coli O157 |
YPJA |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G7382 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG13213 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120003950 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB3005 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0008713 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories
- Genes in OpenBioSystems with Promoter Fusions
- Genes with homologs in Apis mellifera
- Genes with homologs in Bos taurus
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Canis familiaris
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Schizosaccharomyces pombe
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in E. coli O157


