tdcE:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
tdcE |
|---|---|
| Gene Synonym(s) |
ECK3103, b3114, JW5522, yhaS[1], yhaS |
| Product Desc. |
Pyruvate formate-lyase/ketobutyrate formate-lyase[2] |
| Product Synonyms(s) |
pyruvate formate-lyase 4/2-ketobutyrate formate-lyase[1], B3114[3][1], YhaS[3][1], 2-ketobutyrate formate-lyase, active[3][1], pyruvate formate-lyase 4[3][1], PFL4[3][1] , ECK3103, JW5522, yhaS, b3114 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression |
transcription unit(s): tdcABCDEFG[3], OP00167, tdcABC |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
HT_Cmplx9_Cyt: YfiD+TdcE.[2]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
tdcE |
|---|---|
| Mnemonic |
Threonine dehydratase, catabolic |
| Synonyms |
ECK3103, b3114, JW5522, yhaS[1], yhaS |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
70.22 minutes |
MG1655: 3260440..3258146 |
||
|
NC_012967: 3195452..3193158 |
||||
|
NC_012759: 3145294..3147588 |
||||
|
W3110 |
|
W3110: 3261074..3258780 |
||
|
DH10B: 3358185..3355891 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔtdcE (Keio:JW5522) |
deletion |
deletion |
PMID:16738554 |
| |||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW5522 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAAGGTAGATATTGATACCAG Primer 2:CCGAGCGCCTGGGTAAAGGTACG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 16% [5] | ||
|
Linked marker |
est. P1 cotransduction: 1% [5] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G7627 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12758 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120004178 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB2612 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010242 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
TdcE |
|---|---|
| Synonyms |
pyruvate formate-lyase 4/2-ketobutyrate formate-lyase[1], B3114[3][1], YhaS[3][1], 2-ketobutyrate formate-lyase, active[3][1], pyruvate formate-lyase 4[3][1], PFL4[3][1] , ECK3103, JW5522, yhaS, b3114 |
| Product description |
Pyruvate formate-lyase/ketobutyrate formate-lyase[2] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
KETOBUTFORMLY-MONOMER
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0003824 |
catalytic activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001150 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004184 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005949 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0963 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0086 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005975 |
carbohydrate metabolic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005949 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006006 |
glucose metabolic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004184 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008152 |
metabolic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001150 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008415 |
acyltransferase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0012 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008861 |
formate C-acetyltransferase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004184 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008861 |
formate C-acetyltransferase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005949 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016740 |
transferase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0808 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0043875 |
2-ketobutyrate formate-lyase activity |
PMID:9484901 |
IGI: Inferred from Genetic Interaction |
EcoliWiki:pflB
|
F |
complete | ||
|
GO:0005737 |
cytoplasm |
PMID:9484901 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0070689 |
threonine catabolic process to propionate |
PMID:9484901 |
IC: Inferred by Curator |
GO:0043875
|
P |
complete | ||
|
GO:0005624 |
membrane fraction |
PMID:16858726 |
IDA: Inferred from Direct Assay |
C |
complete | |||
| edit table |
Interactions
KETOBUTFORMLY-MONOMER
See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
cytoplasm |
From EcoCyc[6] |
| ||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MKVDIDTSDK LYADAWLGFK GTDWKNEINV RDFIQHNYTP YEGDESFLAE ATPATTELWE KVMEGIRIEN ATHAPVDFDT NIATTITAHD AGYINQPLEK IVGLQTDAPL KRALHPFGGI NMIKSSFHAY GREMDSEFEY LFTDLRKTHN QGVFDVYSPD MLRCRKSGVL TGLPDGYGRG RIIGDYRRVA LYGISYLVRE RELQFADLQS RLEKGEDLEA TIRLREELAE HRHALLQIQE MAAKYGFDIS RPAQNAQEAV QWLYFAYLAA VKSQNGGAMS LGRTASFLDI YIERDFKAGV LNEQQAQELI DHFIMKIRMV RFLRTPEFDS LFSGDPIWAT EVIGGMGLDG RTLVTKNSFR YLHTLHTMGP APEPNLTILW SEELPIAFKK YAAQVSIVTS SLQYENDDLM RTDFNSDDYA IACCVSPMVI GKQMQFFGAR ANLAKTLLYA INGGVDEKLK IQVGPKTAPL MDDVLDYDKV MDSLDHFMDW LAVQYISALN IIHYMHDKYS YEASLMALHD RDVYRTMACG IAGLSVATDS LSAIKYARVK PIRDENGLAV DFEIDGEYPQ YGNNDERVDS IACDLVERFM KKIKALPTYR NAVPTQSILT ITSNVVYGQK TGNTPDGRRA GTPFAPGANP MHGRDRKGAV ASLTSVAKLP FTYAKDGISY TFSIVPAALG KEDPVRKTNL VGLLDGYFHH EADVEGGQHL NVNVMNREML LDAIEHPEKY PNLTIRVSGY AVRFNALTRE QQQDVISRTF TQAL |
| Length |
764 |
| Mol. Wt |
85.937 kDa |
| pI |
5.6 (calculated) |
| Extinction coefficient |
92,140 - 92,765 (calc based on 36 Y, 7 W, and 5 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0010242 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:G7627 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12758 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120004178 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2612 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
88 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
15 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
49 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
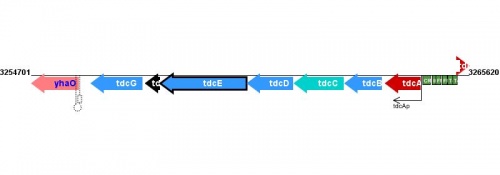 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:3260420..3260460
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for tdcE | |
|
microarray |
Summary of data for tdcE from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to tdcE Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G7627 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2612 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12758 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120004178 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010242 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Shigella flexneri |
TDCE |
From SHIGELLACYC |
|
E. coli O157 |
TDCE |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:G7627 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12758 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120004178 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2612 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010242 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 CGSC: The Coli Genetics Stock Center
- ↑ 5.0 5.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
Categories


