rrlH:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
rrlH |
|---|---|
| Gene Synonym(s) |
ECK0204, b0204, JWR0004[1], JWR0004 |
| Product Desc. |
Component of 50S ribosomal subunit[3] |
| Product Synonyms(s) |
23S rRNA (rrlH)[1], 23S rRNA (rrlG)[1], 23S rRNA (rrlD)[1], 23S rRNA (rrlC)[1], 23S rRNA (rrlA)[1], 23S rRNA (rrlB)[1], 23S rRNA (rrlE)[1], rrlH[2][1], rrnH[2][1] |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression |
transcription unit(s): rrsH-ileV-alaV-rrlH-rrfH[2], rrsH, rrnH |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
rrlH |
|---|---|
| Mnemonic |
Ribosomal RNA, 23S |
| Synonyms |
ECK0204, b0204, JWR0004[1], JWR0004 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
4.87 minutes |
MG1655: 225759..228662 |
||
|
NC_012967: 228588..231493 |
||||
|
NC_012759: 225758..228661 |
||||
|
W3110 |
|
W3110: 225759..228662 |
||
|
DH10B: 199863..202766 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
rrlH MLSr |
A to U base substitution at position 2058 of rrlH |
Resistant to |
Resistance to macrolide-lincosamide-streptogramin |
PMID:3071688 |
See figures 1 to 3 and table 1 for the results of the mapping tests. | ||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
Plasmid clone |
PMID:16769691 Status: Primer 1: Primer 2: | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: % [5] | ||
|
Linked marker |
est. P1 cotransduction: 81% [5] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG30083 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG30083 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120002525 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB4246 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000684 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
23S rRNA |
|---|---|
| Synonyms |
23S rRNA (rrlH)[1], 23S rRNA (rrlG)[1], 23S rRNA (rrlD)[1], 23S rRNA (rrlC)[1], 23S rRNA (rrlA)[1], 23S rRNA (rrlB)[1], 23S rRNA (rrlE)[1], rrlH[2][1], rrnH[2][1] |
| Product description |
Component of 50S ribosomal subunit[3] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of 50S ribosomal subunit |
could be indirect |
| |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
| Type |
rRNA |
|---|---|
| Sequence |
GGUUAAGCGA CUAAGCGUAC ACGGUGGAUG CCCUGGCAGU CAGAGGCGAU GAAGGACGUG CUAAUCUGCG AUAAGCGUCG GUAAGGUGAU AUGAACCGUU AUAACCGGCG AUUUCCGAAU GGGGAAACCC AGUGUGUUUC GACACACUAU CAUUAACUGA AUCCAUAGGU UAAUGAGGCG AACCGGGGGA ACUGAAACAU CUAAGUACCC CGAGGAAAAG AAAUCAACCG AGAUUCCCCC AGUAGCGGCG AGCGAACGGG GAGGAGCCCA GAGCCUGAAU CAGUGUGUGU GUUAGUGGAA GCGUCUGGAA AGGCGCGCGA UACAGGGUGA CAGCCCCGUA CACAAAAAUG CACAUGCUGU GAGCUCGAUG AGUAGGGCGG GACACGUGGU AUCCUGUCUG AAUAUGGGGG GACCAUCCUC CAAGGCUAAA UACUCCUGAC UGACCGAUAG UGAACCAGUA CCGUGAGGGA AAGGCGAAAA GAACCCCGGC GAGGGGAGUG AAAAAGAACC UGAAACCGUG UACGUACAAG CAGUGGGAGC AUGCUUAGGC GUGUGACUGC GUACCUUUUG UAUAAUGGGU CAGCGACUUA UAUUCUGUAG CAAGGUUAAC CGAAUAGGGG AGCCGAAGGG AAACCGAGUC UUAACUGGGC GUUAAGUUGC AGGGUAUAGA CCCGAAACCC GGUGAUCUAG CCAUGGGCAG GUUGAAGGUU GGGUAACACU AACUGGAGGA CCGAACCGAC UAAUGUUGAA AAAUUAGCGG AUGACUUGUG GCUGGGGGUG AAAGGCCAAU CAAACCGGGA GAUAGCUGGU UCUCCCCGAA AGCUAUUUAG GUAGCGCCUC GUGAACUCAU CUCCGGGGGU AGAGCACUGU UUCGGCAAGG GGGUCAUCCC GACUUACCAA CCCGAUGCAA ACUGCGAAUA CCGGAGAAUG UUAUCACGGG AGACACACGG CGGGUGCUAA CGUCCGUCGU GAAGAGGGAA ACAACCCAGA CCGCCAGCUA AGGUCCCAAA GUCAUGGUUA AGUGGGAAAC GAUGUGGGAA GGCCCAGACA GCCAGGAUGU UGGCUUAGAA GCAGCCAUCA UUUAAAGAAA GCGUAAUAGC UCACUGGUCG AGUCGGCCUG CGCGGAAGAU GUAACGGGGC UAAACCAUGC ACCGAAGCUG CGGCAGCGAC GCUUAUGCGU UGUUGGGUAG GGGAGCGUUC UGUAAGCCUG UGAAGGUGUA CUGUGAGGUA UGCUGGAGGU AUCAGAAGUG CGAAUGCUGA CAUAAGUAAC GAUAAAGCGG GUGAAAAGCC CGCUCGCCGG AAGACCAAGG GUUCCUGUCC AACGUUAAUC GGGGCAGGGU GAGUCGACCC CUAAGGCGAG GCCGAAAGGC GUAGUCGAUG GGAAACAGGU UAAUAUUCCU GUACUUGGUG UUACUGCGAA GGGGGGACGG AGAAGGCUAU GUUGGCCGGG CGACGGUUGU CCCGGUUUAA GCGUGUAGGC UGGUUUUCCA GGCAAAUCCG GAAAAUCAAG GCUGAGGCGU GAUGACGAGG CACUACGGUG CUGAAGCAAC AAAUGCCCUG CUUCCAGGAA AAGCCUCUAA GCAUCAGGUA ACAUCAAAUC GUACCCCAAA CCGACACAGG UGGUCAGGUA GAGAAUACCA AGGCGCUUGA GAGAACUCGG GUGAAGGAAC UAGGCAAAAU GGUGCCGUAA CUUCGGGAGA AGGCACGCUG AUAUGUAGGU GAAGCGACUU GCUCGUGGAG CUGAAAUCAG UCGAAGAUAC CAGCUGGCUG CAACUGUUUA UUAAAAACAC AGCACUGUGC AAACACGAAA GUGGACGUAU ACGGUGUGAC GCCUGCCCGG UGCCGGAAGG UUAAUUGAUG GGGUUAGCGC AAGCGAAGCU CUUGAUCGAA GCCCCGGUAA ACGGCGGCCG UAACUAUAAC GGUCCUAAGG UAGCGAAAUU CCUUGUCGGG UAAGUUCCGA CCUGCACGAA UGGCGUAAUG AUGGCCAGGC UGUCUCCACC CGAGACUCAG UGAAAUUGAA CUCGCUGUGA AGAUGCAGUG UACCCGCGGC AAGACGGAAA GACCCCGUGA ACCUUUACUA UAGCUUGACA CUGAACAUUG AGCCUUGAUG UGUAGGAUAG GUGGGAGGCU UUGAAGUGUG GACGCCAGUC UGCAUGGAGC CGACCUUGAA AUACCACCCU UUAAUGUUUG AUGUUCUAAC GUUGACCCGU AAUCCGGGUU GCGGACAGUG UCUGGUGGGU AGUUUGACUG GGGCGGUCUC CUCCUAAAGA GUAACGGAGG AGCACGAAGG UUGGCUAAUC CUGGUCGGAC AUCAGGAGGU UAGUGCAAUG GCAUAAGCCA GCUUGACUGC GAGCGUGACG GCGCGAGCAG GUGCGAAAGC AGGUCAUAGU GAUCCGGUGG UUCUGAAUGG AAGGGCCAUC GCUCAACGGA UAAAAGGUAC UCCGGGGAUA ACAGGCUGAU ACCGCCCAAG AGUUCAUAUC GACGGCGGUG UUUGGCACCU CGAUGUCGGC UCAUCACAUC CUGGGGCUGA AGUAGGUCCC AAGGGUAUGG CUGUUCGCCA UUUAAAGUGG UACGCGAGCU GGGUUUAGAA CGUCGUGAGA CAGUUCGGUC CCUAUCUGCC GUGGGCGCUG GAGAACUGAG GGGGGCUGCU CCUAGUACGA GAGGACCGGA GUGGACGCAU CACUGGUGUU CGGGUUGUCA UGCCAAUGGC ACUGCCCGGU AGCUAAAUGC GGAAGAGAUA AGUGCUGAAA GCAUCUAAGC ACGAAACUUG CCCCGAGAUG AGUUCUCCCU GACUCCUUGA GAGUCCUGAA GGAACGUUGA AGACGACGAC GUUGAUAGGC CGGGUGUGUA AGCGCAGCGA UGCGUUGAGC UAACCGGUAC UAAUGAACCG UGAGGCUUAA CCUU |
| Mol. Wt |
941816.03 |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG30083 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG30083 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120002525 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB4246 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000684 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
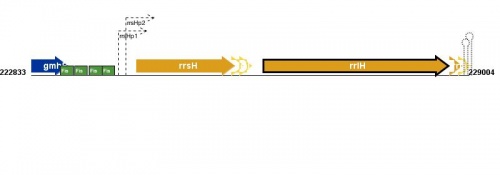 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:225739..225779
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for rrlH | |
|
microarray |
Summary of data for rrlH from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (225502..225801) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to rrlH Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG30083 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB4246 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG30083 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120002525 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000684 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
<protect>
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
| edit table |
</protect>
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
YAED |
From SHIGELLACYC |
|
E. coli O157 |
YAED |
From ECOO157CYC |
| edit table |
<protect></protect>
Notes
Families
<protect>
See Help:Evolution_families for help entering or editing information in this section of EcoliWiki.
| Database | Accession | Notes |
|---|---|---|
| edit table |
</protect> <protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 CGSC: The Coli Genetics Stock Center
- ↑ 5.0 5.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories


