ppc:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
ppc |
|---|---|
| Gene Synonym(s) |
ECK3947, b3956, JW3928, asp, glu, pepC[1] |
| Product Desc. |
Component of phosphoenolpyruvate carboxylase[2][3] Phosphoenolpyruvate carboxylase; monomeric[4] |
| Product Synonyms(s) |
phosphoenolpyruvate carboxylase[1], B3956[2][1], Glu[2][1], Asp[2][1], Ppc[2][1] , asp, ECK3947, glu, JW3928, b3956 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Mutant has reduced growth rate with little acetate excreation, decrease glucose consumption and a decreased carbon dioxide evolution rate.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
ppc |
|---|---|
| Mnemonic |
PEP carboxylase |
| Synonyms |
ECK3947, b3956, JW3928, asp, glu, pepC[1] |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
89.41 minutes, 89.41 minutes |
MG1655: 4151121..4148470 |
||
|
NC_012967: 4133205..4130554 |
||||
|
NC_012759: 4038139..4040790 |
||||
|
W3110 |
|
W3110: 3483583..3486234 |
||
|
DH10B: 4250818..4248167 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ppcR587S |
R587S |
Loss of activity |
seeded from UniProt:P00864 | ||||
|
ppcH579P |
H579P |
5.4% of wild-type activity |
seeded from UniProt:P00864 | ||||
|
ppcH579N |
H579N |
29% of wild-type activity |
seeded from UniProt:P00864 | ||||
|
ppcH138N |
H138N |
Loss of activity |
seeded from UniProt:P00864 | ||||
|
Δppc (Keio:JW3928) |
deletion |
deletion |
PMID:16738554 |
||||
|
Δppc::kan |
deletion |
Biolog:respiration |
unable to respire Dextrin |
PMID:16095938 |
|||
|
Δppc::kan |
deletion |
Biolog:respiration |
unable to respire D-Sorbitol |
PMID:16095938 |
|||
|
Δppc::kan |
deletion |
Biolog:respiration |
unable to respire DL-a-Glycerol |
PMID:16095938 |
|||
|
Δppc::kan |
deletion |
Biolog:respiration |
unable to respire D-Galactoniate |
PMID:16095938 |
|||
|
Δppc::kan |
deletion |
Biolog:respiration |
unable to respire D-Galacturonate |
PMID:16095938 |
|||
|
Δppc::kan |
deletion |
Biolog:respiration |
unable to respire D-Glucuronate |
PMID:16095938 |
|||
|
Δppc::kan |
deletion |
Biolog:respiration |
unable to respire Acetate |
PMID:16095938 |
|||
|
Δppc::kan |
deletion |
Biolog:respiration |
unable to respire a-Hydroxybutyrate |
PMID:16095938 |
|||
|
Δppc::kan |
deletion |
Biolog:respiration |
unable to respire a-Ketobutyrate |
PMID:16095938 |
|||
|
Δppc::kan |
deletion |
Biolog:respiration |
unable to respire Glucuronamide |
PMID:16095938 |
|||
|
Δppc::kan |
deletion |
Biolog:respiration |
unable to respire L-Alanine |
PMID:16095938 |
|||
|
Δppc::kan |
deletion |
Biolog:respiration |
unable to respire L-Serine |
PMID:16095938 |
|||
|
Δppc::kan |
deletion |
Biolog:respiration |
unable to respire Uridine |
PMID:16095938 |
|||
|
ppc-2 |
PMID:13791860 PMID:4874804 PMID:14292146 |
||||||
|
ppc-1 |
PMID:4874804 PMID:14292146 |
||||||
|
ppc-3 |
|||||||
|
ppc-5::kan |
|||||||
|
Δppc-742::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW3928 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAACGAACAATATTCCGCATT Primer 2:CCGCCGGTATTACGCATACCTGC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 84% [6] | ||
|
Linked marker |
est. P1 cotransduction: 88% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10756 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10756 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000747 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0749 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012950 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
Ppc |
|---|---|
| Synonyms |
phosphoenolpyruvate carboxylase[1], B3956[2][1], Glu[2][1], Asp[2][1], Ppc[2][1] , asp, ECK3947, glu, JW3928, b3956 |
| Product description |
Component of phosphoenolpyruvate carboxylase[2][3] Phosphoenolpyruvate carboxylase; monomeric[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0003824 |
catalytic activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR015813 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006099 |
tricarboxylic acid cycle |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00595 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006099 |
tricarboxylic acid cycle |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001449 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006099 |
tricarboxylic acid cycle |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR018129 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006099 |
tricarboxylic acid cycle |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0816 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008964 |
phosphoenolpyruvate carboxylase activity |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00595 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008964 |
phosphoenolpyruvate carboxylase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001449 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008964 |
phosphoenolpyruvate carboxylase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR018129 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008964 |
phosphoenolpyruvate carboxylase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:4.1.1.31 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0015977 |
carbon fixation |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00595 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0015977 |
carbon fixation |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0120 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016829 |
lyase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0456 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of phosphoenolpyruvate carboxylase |
could be indirect |
||
|
Protein |
uvrA |
PMID:16606699 |
Experiment(s):EBI-1147140 | |
|
Protein |
rpmB |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
gcvP |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
glcB |
PMID:19402753 |
LCMS(ID Probability):99.3 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MNEQYSALRS NVSMLGKVLG ETIKDALGEH ILERVETIRK LSKSSRAGND ANRQELLTTL QNLSNDELLP VARAFSQFLN LANTAEQYHS ISPKGEAASN PEVIARTLRK LKNQPELSED TIKKAVESLS LELVLTAHPT EITRRTLIHK MVEVNACLKQ LDNKDIADYE HNQLMRRLRQ LIAQSWHTDE IRKLRPSPVD EAKWGFAVVE NSLWQGVPNY LRELNEQLEE NLGYKLPVEF VPVRFTSWMG GDRDGNPNVT ADITRHVLLL SRWKATDLFL KDIQVLVSEL SMVEATPELL ALVGEEGAAE PYRYLMKNLR SRLMATQAWL EARLKGEELP KPEGLLTQNE ELWEPLYACY QSLQACGMGI IANGDLLDTL RRVKCFGVPL VRIDIRQEST RHTEALGELT RYLGIGDYES WSEADKQAFL IRELNSKRPL LPRNWQPSAE TREVLDTCQV IAEAPQGSIA AYVISMAKTP SDVLAVHLLL KEAGIGFAMP VAPLFETLDD LNNANDVMTQ LLNIDWYRGL IQGKQMVMIG YSDSAKDAGV MAASWAQYQA QDALIKTCEK AGIELTLFHG RGGSIGRGGA PAHAALLSQP PGSLKGGLRV TEQGEMIRFK YGLPEITVSS LSLYTGAILE ANLLPPPEPK ESWRRIMDEL SVISCDVYRG YVRENKDFVP YFRSATPEQE LGKLPLGSRP AKRRPTGGVE SLRAIPWIFA WTQNRLMLPA WLGAGTALQK VVEDGKQSEL EAMCRDWPFF STRLGMLEMV FAKADLWLAE YYDQRLVDKA LWPLGKELRN LQEEDIKVVL AIANDSHLMA DLPWIAESIQ LRNIYTDPLN VLQAELLHRS RQAEKEGQEP DPRVEQALMV TIAGIAAGMR NTG |
| Length |
883 |
| Mol. Wt |
99.065 kDa |
| pI |
5.6 (calculated) |
| Extinction coefficient |
138,770 - 139,770 (calc based on 23 Y, 19 W, and 8 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0012950 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10756 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10756 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000747 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0749 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
8.51E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
14.585+/-0.258 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.180497925 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
2530 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
4441 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
2495 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
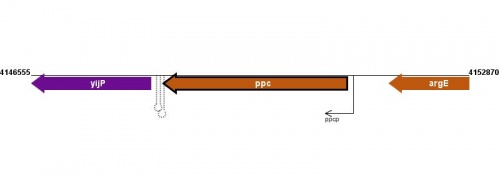 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:4151101..4151141
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for ppc | |
|
microarray |
Summary of data for ppc from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to ppc Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10756 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0749 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10756 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000747 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012950 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
PPC |
From SHIGELLACYC |
|
E. coli O157 |
PPC |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10756 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10756 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000747 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0749 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012950 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories


