hsdR:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
hsdR |
|---|---|
| Gene Synonym(s) |
ECK4340, b4350, JW4313, Hs, hsp, hsr, rm[1] |
| Product Desc. |
host restriction; endonuclease R[2][3]; Component of [[:Category:Complex:EcoKI restriction-modification system|EcoKI restriction-modification system]][3] Endonuclease R, host restriction of foreign DNA; ClpXP-dependent degradation[4] |
| Product Synonyms(s) |
endonuclease R[1], B4350[2][1], Hsr[2][1], Hsp[2][1], HsdR[2][1] , ECK4340, hsp, hsr, JW4313, b4350 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
HsdR acts as a chaperone in hsdS mutant to allow DNA binding.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
hsdR |
|---|---|
| Mnemonic |
Host specificity |
| Synonyms |
ECK4340, b4350, JW4313, Hs, hsp, hsr, rm[1] |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
98.74 minutes |
MG1655: 4584838..4581272 |
||
|
NC_012967: 4564736..4561224 |
||||
|
NC_012759: 4519756..4523322 |
||||
|
W3110 |
|
W3110: 4591495..4587929 |
||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔhsdR (Keio:JW4313) |
deletion |
deletion |
PMID:16738554 |
||||
|
hsdRΔ4 |
deletion |
deletion |
Lacks DNA specifying six DEAD-box motifs that are critical for helicase activity |
PMID: 9776741 |
|||
|
hsdR514 |
|||||||
|
hsdR2 |
|||||||
|
hsdR4 |
|||||||
|
hsdR17 |
|||||||
|
hsdR19 |
|||||||
|
hsdR0 |
|||||||
|
ΔhsdR24 |
|||||||
|
hsdR11 |
|||||||
|
hsdR27 |
|||||||
|
hsdR18 |
|||||||
|
hsdR519 |
|||||||
|
hsdR13 |
|||||||
|
hsdR29 |
|||||||
|
ΔhsdR748::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW4313 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCTTATGGGCgTTAAATATTTGGACAGGCCCG Primer 2:CCGGCCAGCTCGTCCCAGATATA | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: % [6] | ||
|
Linked marker |
est. P1 cotransduction: 66% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10459 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10459 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000452 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0454 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0014264 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
HsdR |
|---|---|
| Synonyms |
endonuclease R[1], B4350[2][1], Hsr[2][1], Hsp[2][1], HsdR[2][1] , ECK4340, hsp, hsr, JW4313, b4350 |
| Product description |
host restriction; endonuclease R[2][3]; Component of [[:Category:Complex:EcoKI restriction-modification system|EcoKI restriction-modification system]][3] Endonuclease R, host restriction of foreign DNA; ClpXP-dependent degradation[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003676 |
nucleic acid binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001650 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006935 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007409 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003677 |
DNA binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0238 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004386 |
helicase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001650 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004386 |
helicase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0347 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004519 |
endonuclease activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007409 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001650 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006935 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006304 |
DNA modification |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007409 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009035 |
Type I site-specific deoxyribonuclease activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:3.1.21.3 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009307 |
DNA restriction-modification system |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0680 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016787 |
hydrolase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006935 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016787 |
hydrolase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0378 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of [[:Category:Complex:EcoKI restriction-modification system|EcoKI restriction-modification system]] |
could be indirect |
||
|
Protein |
aceF |
PMID:15690043 |
Experiment(s):EBI-889458, EBI-894477 | |
|
Protein |
hsdM |
PMID:15690043 |
Experiment(s):EBI-889458, EBI-890115, EBI-894477, EBI-895107 | |
|
Protein |
lpdA |
PMID:15690043 |
Experiment(s):EBI-889458, EBI-894477 | |
|
Protein |
rplL |
PMID:15690043 |
Experiment(s):EBI-894477 | |
|
Protein |
hsdM |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):28.932061 | |
|
Protein |
rplL |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MLWALNIWTG PHSNGLITMM NKSNFEFLKG VNDFTYAIAC AAENNYPDDP NTTLIKMRMF GEATAKHLGL LLNIPPCENQ HDLLRELGKI AFVDDNILSV FHKLRRIGNQ AVHEYHNDLN DAQMCLRLGF RLAVWYYRLV TKDYDFPVPV FVLPERGENL YHQEVLTLKQ QLEQQVREKA QTQAEVEAQQ QKLVALNGYI AILEGKQQET EAQTQARLAA LEAQLAEKNA ELAKQTEQER KAYHKEITDQ AIKRTLNLSE EESRFLIDAQ LRKAGWQADS KTLRFSKGAR PEPGVNKAIA EWPTGKDETG NQGFADYVLF VGLKPIAVVE AKRNNIDVPA RLNESYRYSK CFDNGFLRET LLEHYSPDEV HEAVPEYETS WQDTSGKQRF KIPFCYSTNG REYRATMKTK SGIWYRDVRD TRNMSKALPE WHRPEELLEM LGSEPQKQNQ WFADNPGMSE LGLRYYQEDA VRAVEKAIVK GQQEILLAMA TGTGKTRTAI AMMFRLIQSQ RFKRILFLVD RRSLGEQALG AFEDTRINGD TFNSIFDIKG LTDKFPEDST KIHVATVQSL VKRTLQSDEP MPVARYDCIV VDEAHRGYIL DKEQTEGELQ FRSQLDYVSA YRRILDHFDA VKIALTATPA LHTVQIFGEP VYRYTYRTAV IDGFLIDQDP PIQIITRNAQ EGVYLSKGEQ VERISPQGEV INDTLEDDQD FEVADFNRGL VIPAFNRAVC NELTNYLDPT GSQKTLVFCV TNAHADMVVE ELRAAFKKKY PQLEHDAIIK ITGDADKDAR KVQTMITRFN KERLPNIVVT VDLLTTGVDI PSICNIVFLR KVRSRILYEQ MKGRATRLCP EVNKTSFKIF DCVDIYSTLE SVDTMRPVVV RPKVELQTLV NEITDSETYK ITEADGRSFA EHSHEQLVAK LQRIIGLATF NRDRSETIDK QVRRLDELCQ DAAGVNFNGF ASRLREKGPH WSAEVFNKLP GFIARLEKLK TDINNLNDAP IFLDIDDEVV SVKSLYGDYD TPQDFLEAFD SLVQRSPNAQ PALQAVINRP RDLTRKGLVE LQEWFDRQHF EESSLRKAWK ETRNEDIAAR LIGHIRRAAV GDALKPFEER VDHALTRIKG ENDWSSEQLS WLDRLAQALK EKVVLDDDVF KTGNFHRRGG KAMLQRTFDD NLDTLLGKFS DYIWDELA |
| Length |
1,188 |
| Mol. Wt |
136.103 kDa |
| pI |
5.9 (calculated) |
| Extinction coefficient |
134,650 - 136,150 (calc based on 35 Y, 15 W, and 12 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0014264 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10459 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10459 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000452 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0454 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
1.14E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
E. coli K-12 MG1655 |
404 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
186 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
338 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
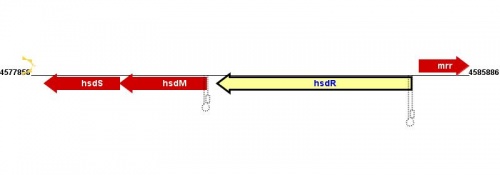 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:4584818..4584858
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for hsdR | |
|
microarray |
Summary of data for hsdR from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (4584295..4584573) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to hsdR Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10459 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0454 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10459 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000452 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0014264 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
<protect>
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
| edit table |
</protect>
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
PF04313 Type I restriction enzyme R protein N terminus (HSDR_N) |
||
|
EcoCyc:EG10459 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10459 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000452 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0454 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0014264 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 3.4 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories


