glgP:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
glgP |
|---|---|
| Gene Synonym(s) |
ECK3414, b3428, JW3391, glgY[1], glgY |
| Product Desc. |
Component of glycogen phosphorylase[2][3] Glycogen phosphorylase; alpha-glucan phosphorylase[4] |
| Product Synonyms(s) |
glycogen phosphorylase[1], B3428[2][1], GlgY[2][1], GlgP[2][1] , ECK3414, glgY, JW3391, b3428 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
glgP |
|---|---|
| Mnemonic |
Glycogen |
| Synonyms |
ECK3414, b3428, JW3391, glgY[1], glgY |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
76.78 minutes |
MG1655: 3564604..3562157 |
||
|
NC_012967: 3498035..3495588 |
||||
|
NC_012759: 3450651..3453098 |
||||
|
W3110 |
|
W3110: 4073834..4076281 |
||
|
DH10B: 3662349..3659902 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔglgP (Keio:JW3391) |
deletion |
deletion |
PMID:16738554 |
||||
|
glgP721::Tn10 |
Insertion at 3,564,565 bp in MG1655 (NC_000913) |
adapted from Nichols et al.[6] |
Synonyms: zhg-3086::Tn10, zhh-21::Tn10 | ||||
|
glgP721::Tn10 |
PMID:3540312 PMID:9829956 |
||||||
|
ΔglgP761::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW3391 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAATGCTCCGTTTACATATTC Primer 2:CCtAATCTCACCGGATCGATATG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 5% [6] | ||
|
zhg-50::Tn10 |
Linked marker |
est. P1 cotransduction: 33% [6] | |
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10380 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10380 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000373 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0375 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0011198 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
GlgP |
|---|---|
| Synonyms |
glycogen phosphorylase[1], B3428[2][1], GlgY[2][1], GlgP[2][1] , ECK3414, glgY, JW3391, b3428 |
| Product description |
Component of glycogen phosphorylase[2][3] Glycogen phosphorylase; alpha-glucan phosphorylase[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0016052 |
carbohydrate catabolic process |
PMID:4878695 |
IDA: Inferred from Direct Assay |
P |
Seeded from Riley et al 2006 [1]. |
complete | ||
|
GO:0000271 |
polysaccharide biosynthetic process |
P |
Seeded from Riley et al 2006 [1]. |
Missing: evidence, reference | ||||
|
GO:0030170 |
pyridoxal phosphate binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011833 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of glycogen phosphorylase |
could be indirect |
||
|
Protein |
intA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
pykF |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
rpmB |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
ptsH |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MNAPFTYSSP TLSVEALKHS IAYKLMFTIG KDPVVANKHE WLNATLFAVR DRLVERWLRS NRAQLSQETR QVYYLSMEFL IGRTLSNAML SLGIYEDVQG ALEAMGLNLE ELIDEENDPG LGNGGLGRLA ACFLDSLATL GLPGRGYGIR YDYGMFKQNI VNGSQKESPD YWLEYGNPWE FKRHNTRYKV RFGGRIQQEG KKTRWIETEE ILGVAYDQII PGYDTDATNT LRLWSAQASS EINLGKFNQG DYFAAVEDKN HSENVSRVLY PDDSTYSGRE LRLRQEYFLV SSTIQDILSR HYQLHKTYDN LADKIAIHLN DTHPVLSIPE MMRLLIDEHQ FSWDDAFEVC CQVFSYTNHT LMSEALETWP VDMLGKILPR HLQIIFEIND YFLKTLQEQY PNDTDLLGRA SIIDESNGRR VRMAWLAVVV SHKVNGVSEL HSNLMVQSLF ADFAKIFPGR FTNVTNGVTP RRWLAVANPS LSAVLDEHLG RNWRTDLSLL NELQQHCDFP MVNHAVHQAK LENKKRLAEY IAQQLNVVVN PKALFDVQIK RIHEYKRQLM NVLHVITRYN RIKADPDAKW VPRVNIFGGK AASAYYMAKH IIHLINDVAK VINNDPQIGD KLKVVFIPNY SVSLAQLIIP AADLSEQISL AGTEASGTSN MKFALNGALT IGTLDGANVE MLDHVGADNI FIFGNTAEEV EELRRQGYKP REYYEKDEEL HQVLTQIGSG VFSPEDPGRY RDLVDSLINF GDHYQVLADY RSYVDCQDKV DELYELQEEW TAKAMLNIAN MGYFSSDRTI KEYADHIWHI DPVRL |
| Length |
815 |
| Mol. Wt |
93.174 kDa |
| pI |
5.9 (calculated) |
| Extinction coefficient |
133,620 - 134,245 (calc based on 38 Y, 14 W, and 5 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0011198 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10380 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10380 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000373 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0375 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
Ecoli K-12 |
57.187+/-0.397 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.16591745 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
178 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
427 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
177 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
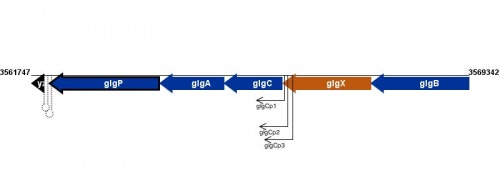 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:3564584..3564624
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for glgP | |
|
microarray |
Summary of data for glgP from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to glgP Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10380 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0375 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10380 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000373 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0011198 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Danio rerio |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Drosophila melanogaster |
|
From Inparanoid:20070104 |
|
Drosophila pseudoobscura |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Macaca mulatta |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Saccharomyces cerevisiae |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Tetraodon nigroviridis |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
GLGP |
From SHIGELLACYC |
|
E. coli O157 |
GLGP |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10380 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10380 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000373 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0375 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0011198 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 5.3 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 6.2 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Apis mellifera
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Bos taurus
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Canis familiaris
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Danio rerio
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Drosophila melanogaster
- Genes with homologs in Drosophila pseudoobscura
- Genes with homologs in Gallus gallus
- Genes with homologs in Homo sapiens
- Genes with homologs in Macaca mulatta
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Oryza gramene
- Genes with homologs in Pan troglodytes
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Saccharomyces cerevisiae
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Tetraodon nigroviridis
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


