evgS:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
evgS |
|---|---|
| Gene Synonym(s) |
ECK2366, b2370, JW2367[1], JW2367 |
| Product Desc. |
Sensor kinase for acid and drug resistance, cognate to EvgA[4] |
| Product Synonyms(s) |
hybrid sensory histidine kinase in two-component regulatory system with EvgA [1], B2370 [2][1], sensor kinase-phosphotransferase EvgS-Phis[2][1], phospho-EvgS[2][1], phosphorylated sensor kinase-phosphotransferase EvgS[2][1], modified sensor kinase-phosphotransferase EvgS t[2][1] , ECK2366, JW2367, b2370 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Overexpression causes abnormal biofilm architecture. First 21 aa are predicted to be a type I signal peptide.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
evgS |
|---|---|
| Mnemonic |
E. coli homolog of Virulence Gene |
| Synonyms |
ECK2366, b2370, JW2367[1], JW2367 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
53.5 minutes |
MG1655: 2482396..2485989 |
||
|
NC_012967: 2418601..2422194 |
||||
|
NC_012759: 2368201..2371794 |
||||
|
W3110 |
|
W3110: 2489820..2493413 |
||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
Strain DH10B |
L152F |
Nonsynonomous mutation |
PMID:18245285 |
This is a SNP in E. coli K-12 Strain DH10B | |||
|
ΔevgS (Keio:JW2367) |
deletion |
deletion |
PMID:16738554 |
||||
|
evgS::Tn5KAN-2 (FB20779) |
Insertion at nt 3112 in Plus orientation |
PMID:15262929 |
contains pKD46 | ||||
|
evgSF577S |
F577S |
(in EvgS1; constitutively active) |
Strain variation; seeded from UniProt:P30855 | ||||
|
evgSE701G |
E701G |
(in EvgS4; constitutively active) |
Strain variation; seeded from UniProt:P30855 | ||||
|
ΔevgS779::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW2367 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCAAGTTTTTACCCTATATTTT Primer 2:CCGTCATTTTTCTGACAGAAAAC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: % [6] | ||
|
zfd-1::Tn10 |
Linked marker |
est. P1 cotransduction: 73% [6] | |
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11610 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11610 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001558 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB1567 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0007819 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
EvgS |
|---|---|
| Synonyms |
hybrid sensory histidine kinase in two-component regulatory system with EvgA [1], B2370 [2][1], sensor kinase-phosphotransferase EvgS-Phis[2][1], phospho-EvgS[2][1], phosphorylated sensor kinase-phosphotransferase EvgS[2][1], modified sensor kinase-phosphotransferase EvgS t[2][1] , ECK2366, JW2367, b2370 |
| Product description |
Sensor kinase for acid and drug resistance, cognate to EvgA[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0000155 |
two-component sensor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003661 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000155 |
two-component sensor activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005467 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000156 |
two-component response regulator activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001789 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000160 |
two-component signal transduction system (phosphorelay) |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001789 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000160 |
two-component signal transduction system (phosphorelay) |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008207 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000160 |
two-component signal transduction system (phosphorelay) |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0902 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0000166 |
nucleotide binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0547 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004673 |
protein histidine kinase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005467 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004673 |
protein histidine kinase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:2.7.13.3 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004871 |
signal transducer activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR008207 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004871 |
signal transducer activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR009082 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005215 |
transporter activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001638 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003594 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005524 |
ATP binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0067 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0997 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-1003 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006355 |
regulation of transcription, DNA-dependent |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001789 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006810 |
transport |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001638 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003661 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005467 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0007165 |
signal transduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR009082 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR003661 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0472 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0812 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-9909 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016301 |
kinase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0418 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016310 |
phosphorylation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004358 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016740 |
transferase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0808 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016772 |
transferase activity, transferring phosphorus-containing groups |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004358 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0018106 |
peptidyl-histidine phosphorylation |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR005467 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030288 |
outer membrane-bounded periplasmic space |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001638 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
dnaE |
PMID:16606699 |
Experiment(s):EBI-1142662 | |
| edit table |
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
plasma membrane |
From EcoCyc[3] |
| ||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MKFLPYIFLL CCGLWSTISF ADEDYIEYRG ISSNNRVTLD PLRLSNKELR WLASKKNLVI AVHKSQTATL LHTDSQQRVR GINADYLNLL KRALNIKLTL REYADHQKAM DALAEGEVDI VLSHLVTSPP LNNDIAATKP LIITFPALVT TLHDSMRPLT SPKPVNIARV ANYPPDEVIH QSFPKATIIS FTNLYQALAS VSAGHNDYFI GSNIITSSMI SRYFTHSLNV VKYYNSPRQY NFFLTRKESV ILNEVLNRFV DALTNEVRYE VSQNWLDTGN LAFLNKPLEL TEHEKQWIKQ HPNLKVLENP YSPPYSMTDE NGSVRGVMGD ILNIITLQTG LNFSPITVSH NIHAGTQLSP GGWDIIPGAI YSEDRENNVL FAEAFITTPY VFVMQKAPDS EQTLKKGMKV AIPYYYELHS QLKEMYPEVE WIQVDNASAA FHKVKEGELD ALVATQLNSR YMIDHYYPNE LYHFLIPGVP NASLSFAFPR GEPELKDIIN KALNAIPPSE VLRLTEKWIK MPNVTIDTWD LYSEQFYIVT TLSVLLVGSS LLWGFYLLRS VRRRKVIQGD LENQISFRKA LSDSLPNPTY VVNWQGNVIS HNSAFEHYFT ADYYKNAMLP LENSDSPFKD VFSNAHEVTA ETKENRTIYT QVFEIDNGIE KRCINHWHTL CNLPASDNAV YICGWQDITE TRDLINALEV EKNKAIKATV AKSQFLATMS HEIRTPISSI MGFLELLSGS GLSKEQRVEA ISLAYATGQS LLGLIGEILD VDKIESGNYQ LQPQWVDIPT LVQNTCHSFG AIAASKSIAL SCSSTFPEHY LVKIDPQAFK QVLSNLLSNA LKFTTEGAVK ITTSLGHIDD NHAVIKMTIM DSGSGLSQEE QQQLFKRYSQ TSAGRQQTGS GLGLMICKEL IKNMQGDLSL ESHPGIGTTF TITIPVEISQ QVATVEAKAE QPITLPEKLS ILIADDHPTN RLLLKRQLNL LGYDVDEATD GVQALHKVSM QHYDLLITDV NMPNMDGFEL TRKLREQNSS LPIWGLTANA QANEREKGLS CGMNLCLFKP LTLDVLKTHL SQLHQVAHIA PQYRHLDIEA LKNNTANDLQ LMQEILMTFQ HETHKDLPAA FQALEAGDNR TFHQCIHRIH GAANILNLQK LINISHQLEI TPVSDDSKPE ILQLLNSVKE HIAELDQEIA VFCQKND |
| Length |
1,197 |
| Mol. Wt |
134.746 kDa |
| pI |
6.2 (calculated) |
| Extinction coefficient |
138,090 - 139,590 (calc based on 41 Y, 14 W, and 12 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0007819 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG11610 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11610 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120001558 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1567 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
8.16E+01 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
5.08+/-0.419 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.018369051 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
35 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
69 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
20 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
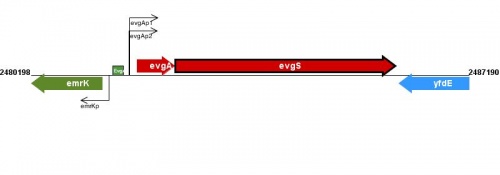 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:2482376..2482416
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for evgS | |
|
microarray |
Summary of data for evgS from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to evgS Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11610 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1567 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11610 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001558 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0007819 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
EVGS |
From SHIGELLACYC |
|
E. coli O157 |
EVGS |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
PF00497 Bacterial extracellular solute-binding proteins, family 3 |
||
|
PF02518 Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase |
||
|
PF00497 Bacterial extracellular solute-binding proteins, family 3 |
||
|
EcoCyc:EG11610 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11610 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001558 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1567 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0007819 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories


