emrB:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
emrB |
|---|---|
| Gene Synonym(s) |
ECK2680, b2686, JW2661[1], JW2661 |
| Product Desc. |
EmrB multidrug MFS transporter[2][3]; Component of EmrAB-TolC Multidrug Efflux Transport System[3] EmrAB-TolC multidrug resistance pump; energy-dependent efflux of octane, thiolactomycin, and uncouplers DNP, CCCP, and FCCP; inner membrane-spanning translocase[4] |
| Product Synonyms(s) |
multidrug efflux system protein[1], B2686[2][1], EmrB[2][1] , ECK2680, JW2661, b2686 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Major Facilitator Superfamily (MFS).[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
emrB |
|---|---|
| Mnemonic |
Efflux-multidrug resistance |
| Synonyms |
ECK2680, b2686, JW2661[1], JW2661 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
60.58 minutes |
MG1655: 2810638..2812176 |
||
|
NC_012967: 2707475..2709013 |
||||
|
NC_012759: 2696450..2697988 |
||||
|
W3110 |
|
W3110: 2811272..2812810 |
||
|
DH10B: 2903180..2904718 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔemrB (Keio:JW2661) |
deletion |
deletion |
PMID:16738554 |
||||
|
ΔemrB767::kan |
PMID:16738554 |
||||||
|
emrB in mutant strain CDM5 |
Resistant to |
Resistant to Thiolactomycin in vivo. It is still sensitive in Vitro. |
PMID:8509326 |
Strain: CDM5 Parental Strain: UB1005 |
See figure 2. | ||
|
pEMR2.1 in strain RP437 |
Resistant to |
This milticopy plasmid in strain RP437, emrA + & emrB + in pUC18, is resistant to Nalidixic acid |
PMID:1409590 |
Parental: RP437 CGSC Expiramental: RP437 with plasmid pEMR2.1 |
|||
|
pEMR2.1 in strain RP437 |
Resistant to |
This milticopy plasmid in strain RP437, emrA + & emrB + in pUC18, is resistant to Carbonyl cyanide m-chlorophenyl hydrazone (CCCP). |
PMID:1409590 |
Parental: RP437 CGSC Expiramental: RP437 with plasmid pEMR2.1 |
|||
|
emrB::Tn10(Kan) |
Resistant to |
Resistance to Thiolactomycin |
PMID:8509326 |
Experimental Strain: SJ261 Parental Strain: UB1005 |
See table 2 | ||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW2661 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCCAACAGCAAAAACCGCTGGA Primer 2:CCGTGCGCACCGCCTCCGCCGCC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 1% [6] | ||
|
Linked marker |
est. P1 cotransduction: 59% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11439 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11439 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001403 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB1409 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0008840 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
EmrB |
|---|---|
| Synonyms |
multidrug efflux system protein[1], B2686[2][1], EmrB[2][1] , ECK2680, JW2661, b2686 |
| Product description |
EmrB multidrug MFS transporter[2][3]; Component of EmrAB-TolC Multidrug Efflux Transport System[3] EmrAB-TolC multidrug resistance pump; energy-dependent efflux of octane, thiolactomycin, and uncouplers DNP, CCCP, and FCCP; inner membrane-spanning translocase[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0997 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-1003 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006810 |
transport |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004638 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006810 |
transport |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0813 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0472 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR004638 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0812 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-9909 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046677 |
response to antibiotic |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0046 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055085 |
transmembrane transport |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011701 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of EmrAB-TolC Multidrug Efflux Transport System |
could be indirect |
| |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
plasma membrane |
C-terminus localized in the cytoplasm with 12 predicted transmembrane domains |
Daley et al. (2005) [7] |
||
|
plasma membrane |
From EcoCyc[3] |
| ||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MQQQKPLEGA QLVIMTIALS LATFMQVLDS TIANVAIPTI AGNLGSSLSQ GTWVITSFGV ANAISIPLTG WLAKRVGEVK LFLWSTIAFA IASWACGVSS SLNMLIFFRV IQGIVAGPLI PLSQSLLLNN YPPAKRSIAL ALWSMTVIVA PICGPILGGY ISDNYHWGWI FFINVPIGVA VVLMTLQTLR GRETRTERRR IDAVGLALLV IGIGSLQIML DRGKELDWFS SQEIIILTVV AVVAICFLIV WELTDDNPIV DLSLFKSRNF TIGCLCISLA YMLYFGAIVL LPQLLQEVYG YTATWAGLAS APVGIIPVIL SPIIGRFAHK LDMRRLVTFS FIMYAVCFYW RAYTFEPGMD FGASAWPQFI QGFAVACFFM PLTTITLSGL PPERLAAASS LSNFTRTLAG SIGTSITTTM WTNRESMHHA QLTESVNPFN PNAQAMYSQL EGLGMTQQQA SGWIAQQITN QGLIISANEI FWMSAGIFLV LLGLVWFAKP PFGAGGGGGG AH |
| Length |
512 |
| Mol. Wt |
55.626 kDa |
| pI |
8.1 (calculated) |
| Extinction coefficient |
104,390 - 105,265 (calc based on 11 Y, 16 W, and 7 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0008840 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG11439 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11439 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120001403 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1409 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
181 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
40 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
59 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
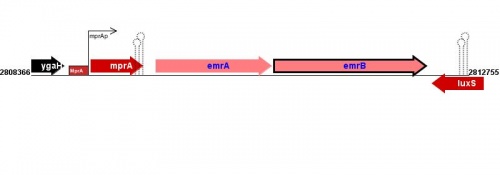 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:2810618..2810658
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for emrB | |
|
microarray |
Summary of data for emrB from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to emrB Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11439 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1409 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11439 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001403 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0008840 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Saccharomyces cerevisiae |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
EMRB |
From SHIGELLACYC |
|
E. coli O157 |
EMRB |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11439 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11439 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001403 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1409 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0008840 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 3.4 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Daley, DO et al. (2005) Global topology analysis of the Escherichia coli inner membrane proteome. Science 308 1321-3 PubMed
Categories
- TMHMM Prediction
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Saccharomyces cerevisiae
- Genes with homologs in Schizosaccharomyces pombe
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157



