add:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
add |
|---|---|
| Gene Synonym(s) |
ECK1618, b1623, JW1615[1], JW1615 |
| Product Desc. |
Adenosine deaminase; mutants affect growth on deoxyadenosine in purA, B mutants[2] |
| Product Synonyms(s) |
adenosine deaminase[1], B1623[3][1], Add[3][1] , ECK1618, JW1615, b1623 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Adenosine deaminase is an enzyme involved in purine metabolism, which irreversibly deaminates both adenosine and deoxyadenosine to inosine and deoxyinosine. The optimal pH for this enzyme is around 7.5-8, but activity is irreversibly lost when pH is lower than 2.7 or higher than 9.5. Adenosine deaminase is sensitive to heavy metals, such as Ag+, Cu2+, and Cr3+, and reacts with them irreversibly (Koch, 1958). [4]
Complications in purine metabolism often result in various disorders in humans. Deficiencies amounts of adenosine deaminase result in diseases such as severe combined immunodeficiency disorder (SCID) and lymphopenia.
Binds TrxA (Kumar, 2004).[2]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
add |
|---|---|
| Mnemonic |
Adenosine deaminase |
| Synonyms |
ECK1618, b1623, JW1615[1], JW1615 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
36.65 minutes |
MG1655: 1700257..1701258 |
||
|
NC_012967: 1679427..1680428 |
||||
|
NC_012759: 1592316..1593317 |
||||
|
W3110 |
|
W3110: 1703947..1704948 |
||
|
DH10B: 1791218..1792219 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
Δadd (Keio:JW1615) |
deletion |
deletion |
PMID:16738554 |
||||
|
add-6 |
|||||||
|
Δadd-771::kan |
PMID:16738554 |
||||||
|
S Φ 3834 |
Tet resistant |
PMID:1998686 |
rpsL, Δadd-uid-man, metB, guaA, uraA::TnlO | ||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW1615 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCATTGATACCACCCTGCCATT Primer 2:CCtTTCGCGGCGACTTTTTCTCG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: % [6] | ||
|
zdi-925::Tn10 |
Linked marker |
est. P1 cotransduction: 1% [6] | |
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10030 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10030 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000027 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0029 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0005435 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
Add |
|---|---|
| Synonyms |
adenosine deaminase[1], B1623[3][1], Add[3][1] , ECK1618, JW1615, b1623 |
| Product description |
Adenosine deaminase; mutants affect growth on deoxyadenosine in purA, B mutants[2] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Adenosine deaminase is the enzyme that catalyzes the irreversible deamination of adenosine or deoxyadenosine to inosine or deoxyinosine. The active site of ADA contains residues of a Histidine, 2 Aspartic acids, one Glutamic acid, and a zinc atom. The overall mechanism involves a hydroxyl group addition to C6 of adenosine, which forms the tetrahedral transition state intermediate. Then, the ammonia elimination forms inosine. Zinc acts as a powerful electrophile, which allows a water molecule to attack C6. Glutamic acid interacts with N1 and donates a proton, reducing the N1=C6 double bond character, allowing the hydroxyl nucleophilic attack. A Histidine residue and the carboxylic group of aspartic acid stabilizes and orients the zinc-bound water molecule. The ammonia elimination pathway is not quite understood but is the final step in the mechanism of adenosine deaminase.
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0005737 |
cytoplasm |
C |
Seeded from Riley et al 2006 [1]. |
Missing: evidence, reference | ||||
|
GO:0004000 |
adenosine deaminase activity |
PMID:4874315 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0004000 |
adenosine deaminase activity |
PMID:765747 |
IMP: Inferred from Mutant Phenotype |
F |
complete | |||
|
GO:0015950 |
purine nucleotide interconversion |
PMID:1998686 |
IGI: Inferred from Genetic Interaction |
EcoliWiki:guaA
|
P |
complete | ||
|
GO:0004000 |
adenosine deaminase activity |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00540 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009168 |
purine ribonucleoside monophosphate biosynthetic process |
PMID:1998686 |
IGI: Inferred from Genetic Interaction |
EcoliWiki:guaA
|
P |
complete | ||
|
GO:0004000 |
adenosine deaminase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006330 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005488 |
binding |
PMID:3486673 |
ISS: Inferred from Sequence or Structural Similarity |
UniProtKB:P56658 |
F |
CHEBI:27834 (Deoxycoformycin mechanistically inhibits Add). |
complete | |
|
GO:0006154 |
adenosine catabolic process |
PMID:1998686 |
IGI: Inferred from Genetic Interaction |
EcoliWiki:guaA
|
P |
complete | ||
|
GO:0004000 |
adenosine deaminase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:3.5.4.4 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004000 |
adenosine deaminase activity |
PMID:5335916 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0032261 |
purine nucleotide salvage |
PMID:1998686 |
IGI: Inferred from Genetic Interaction |
EcoliWiki:guaA
|
P |
complete | ||
|
GO:0006154 |
adenosine catabolic process |
PMID:1998686 |
IGI: Inferred from Genetic Interaction |
P |
Seeded from EcoCyc (v14.0) |
Missing: with/from | ||
|
GO:0043103 |
hypoxanthine salvage |
PMID:1998686 |
IGI: Inferred from Genetic Interaction |
EcoliWiki:guaA
|
P |
complete | ||
|
GO:0009117 |
nucleotide metabolic process |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0546 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046101 |
hypoxanthine biosynthetic process |
PMID:1998686 |
IGI: Inferred from Genetic Interaction |
EcoliWiki:guaA
|
P |
complete | ||
|
GO:0009168 |
purine ribonucleoside monophosphate biosynthetic process |
GOA:hamap |
IEA: Inferred from Electronic Annotation |
HAMAP:MF_00540 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
PMID:5335916 |
IC: Inferred by Curator |
C |
Missing: with/from | |||
|
GO:0009168 |
purine ribonucleoside monophosphate biosynthetic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001365 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009168 |
purine ribonucleoside monophosphate biosynthetic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006330 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009168 |
purine ribonucleoside monophosphate biosynthetic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006650 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009168 |
purine ribonucleoside monophosphate biosynthetic process |
PMID:1998686 |
IGI: Inferred from Genetic Interaction |
P |
Seeded from EcoCyc (v14.0) |
Missing: with/from | ||
|
GO:0015950 |
purine nucleotide interconversion |
PMID:1998686 |
IGI: Inferred from Genetic Interaction |
P |
Seeded from EcoCyc (v14.0) |
Missing: with/from | ||
|
GO:0032261 |
purine nucleotide salvage |
PMID:1998686 |
IGI: Inferred from Genetic Interaction |
P |
Seeded from EcoCyc (v14.0) |
Missing: with/from | ||
|
GO:0043103 |
hypoxanthine salvage |
PMID:1998686 |
IGI: Inferred from Genetic Interaction |
P |
Seeded from EcoCyc (v14.0) |
Missing: with/from | ||
|
GO:0006974 |
response to DNA damage stimulus |
PMID:11967071 |
IEP: Inferred from Expression Pattern |
P |
complete | |||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
rpsM |
PMID:16606699 |
Experiment(s):EBI-1140315 | |
|
Protein |
arpA |
PMID:16606699 |
Experiment(s):EBI-1140315 | |
|
Protein |
rplT |
PMID:16606699 |
Experiment(s):EBI-1140315 | |
|
Protein |
insC1 |
PMID:16606699 |
Experiment(s):EBI-1140315 | |
|
Protein |
ligA |
PMID:16606699 |
Experiment(s):EBI-1140315 | |
|
Protein |
gcd |
PMID:16606699 |
Experiment(s):EBI-1140315 | |
|
Small Molecule |
CHEBI:27834 (Deoxycoformycin) |
Inhibitor of Add activity |
| |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MIDTTLPLTD IHRHLDGNIR PQTILELGRQ YNISLPAQSL ETLIPHVQVI ANEPDLVSFL TKLDWGVKVL ASLDACRRVA FENIEDAARH GLHYVELRFS PGYMAMAHQL PVAGVVEAVI DGVREGCRTF GVQAKLIGIM SRTFGEAACQ QELEAFLAHR DQITALDLAG DELGFPGSLF LSHFNRARDA GWHITVHAGE AAGPESIWQA IRELGAERIG HGVKAIEDRA LMDFLAEQQI GIESCLTSNI QTSTVAELAA HPLKTFLEHG IRASINTDDP GVQGVDIIHE YTVAAPAAGL SREQIRQAQI NGLEMAFLSA EEKRALREKV AAK |
| Length |
333 |
| Mol. Wt |
36.397 kDa |
| pI |
5.4 (calculated) |
| Extinction coefficient |
22,460 - 22,960 (calc based on 4 Y, 3 W, and 4 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0005435 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10030 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10030 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000027 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0029 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
2.89E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
E. coli K-12 MG1655 |
724 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
265 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
548 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
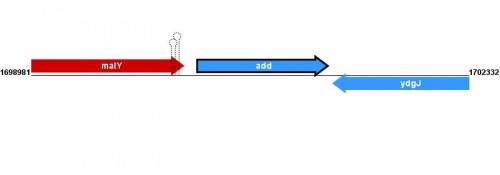 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:1700237..1700277
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for add | |
|
microarray |
Summary of data for add from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (1700057..1700309) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to add Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10030 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0029 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10030 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000027 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0005435 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Danio rerio |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Drosophila melanogaster |
|
From Inparanoid:20070104 |
|
Drosophila pseudoobscura |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Macaca mulatta |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Saccharomyces cerevisiae |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Tetraodon nigroviridis |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
ADD |
From SHIGELLACYC |
|
E. coli O157 |
ADD |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10030 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10030 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000027 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0029 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0005435 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 3.0 3.1 3.2 3.3 3.4 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ KOCH, AL & VALLEE, G (1959) The properties of adenosine deaminase and adenosine nucleoside phosphorylase in extracts of Escherichia coli. J. Biol. Chem. 234 1213-8 PubMed
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories
- Genes in OpenBioSystems with Promoter Fusions
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Apis mellifera
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Bos taurus
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Canis familiaris
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Danio rerio
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Drosophila melanogaster
- Genes with homologs in Drosophila pseudoobscura
- Genes with homologs in Gallus gallus
- Genes with homologs in Homo sapiens
- Genes with homologs in Macaca mulatta
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Pan troglodytes
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Saccharomyces cerevisiae
- Genes with homologs in Schizosaccharomyces pombe
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Tetraodon nigroviridis
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


