yjeP:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
yjeP |
|---|---|
| Gene Synonym(s) |
ECK4155, b4159, JW4120[1], JW4120 |
| Product Desc. |
YjeP : Small Mechanosensitive Ion Channel (MscS) Homolog[2]; predicted mechanosensitive channel[3] Putative mechanosensitive channel protein, function unknown; deletion mutant has no obvious phenotype[4] |
| Product Synonyms(s) |
predicted mechanosensitive channel[1], B4159[2][1], YjeP[2][1] , ECK4155, JW4120, b4159 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
mscS, kefA, yjeP, ybiO, ybdG and ynaI are paralogs.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
yjeP |
|---|---|
| Mnemonic |
Systematic nomenclature |
| Synonyms |
ECK4155, b4159, JW4120[1], JW4120 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
94.49 minutes |
MG1655: 4387393..4384070 |
||
|
NC_012967: 4367525..4364202 |
||||
|
NC_012759: 4322805..4326128 |
||||
|
W3110 |
|
W3110: 4394050..4390727 |
||
|
DH10B: 4487755..4484432 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔyjeP (Keio:JW4120) |
deletion |
deletion |
PMID:16738554 |
||||
|
ΔyjeP786::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW4120 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCCGCCTGATTATCACTTTTCT Primer 2:CCtAAACTTCCCGCCTGACGACC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 35% [6] | ||
|
Linked marker |
est. P1 cotransduction: 16% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB: |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE: |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP: |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
YjeP |
|---|---|
| Synonyms |
predicted mechanosensitive channel[1], B4159[2][1], YjeP[2][1] , ECK4155, JW4120, b4159 |
| Product description |
YjeP : Small Mechanosensitive Ion Channel (MscS) Homolog[2]; predicted mechanosensitive channel[3] Putative mechanosensitive channel protein, function unknown; deletion mutant has no obvious phenotype[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-1003 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006685 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006686 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011014 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011066 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0472 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0812 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-9909 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055085 |
transmembrane transport |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006685 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
lon |
PMID:16606699 |
Experiment(s):EBI-1147585 | |
|
Protein |
dnaE |
PMID:16606699 |
Experiment(s):EBI-1147585 | |
|
Protein |
clpA |
PMID:16606699 |
Experiment(s):EBI-1147585 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MRLIITFLMA WCLSWGAYAA TAPDSKQITQ ELEQAKAAKP AQPEVVEALQ SALNALEERK GSLERIKQYQ QVIDNYPKLS ATLRAQLNNM RDEPRSVSPG MSTDALNQEI LQVSSQLLDK SRQAQQEQER AREIADSLNQ LPQQQTDARR QLNEIERRLG TLTGNTPLNQ AQNFALQSDS ARLKALVDEL ELAQLSANNR QELARLRSEL AEKESQQLDA YLQALRNQLN SQRQLEAERA LESTELLAEN SADLPKDIVA QFKINRELSA ALNQQAQRMD LVASQQRQAA SQTLQVRQAL NTLREQSQWL GSSNLLGEAL RAQVARLPEM PKPQQLDTEM AQLRVQRLRY EDLLNKQPLL RQIHQADGQP LTAEQNRILE AQLRTQRELL NSLLQGGDTL LLELTKLKVS NGQLEDALKE VNEATHRYLF WTSDVRPMTI AWPLEIAQDL RRLISLDTFS QLGKASVMML TSKETILPLF GALILVGCSI YSRRYFTRFL ERSAAKVGKV TQDHFWLTLR TLFWSILVAS PLPVLWMTLG YGLREAWPYP LAVAIGDGVT ATVPLLWVVM ICATFARPNG LFIAHFGWPR ERVSRGMRYY LMSIGLIVPL IMALMMFDNL DDREFSGSLG RLCFILICGA LAVVTLSLKK AGIPLYLNKE GSGDNITNHM LWNMMIGAPL VAILASAVGY LATAQALLAR LETSVAIWFL LLVVYHVIRR WMLIQRRRLA FDRAKHRRAE MLAQRARGEE EAHHHSSPEG AIEVDESEVD LDAISAQSLR LVRSILMLIA LLSVIVLWSE IHSAFGFLEN ISLWDVTSTV QGVESLEPIT LGAVLIAILV FIITTQLVRN LPALLELAIL QHLDLTPGTG YAITTITKYL LMLIGGLVGF SMIGIEWSKL QWLVAALGVG LGFGLQEIFA NFISGLIILF EKPIRIGDTV TIRDLTGSVT KINTRATTIS DWDRKEIIVP NKAFITEQFI NWSLSDSVTR VVLTIPAPAD ANSEEVTEIL LTAARRCSLV IDNPAPEVFL VDLQQGIQIF ELRIYAAEMG HRMPLRHEIH QLILAGFHAH GIDMPFPPFQ MRLESLNGKQ TGRTLTSAGK GRQAGSL |
| Length |
1,107 |
| Mol. Wt |
123.972 kDa |
| pI |
10.5 (calculated) |
| Extinction coefficient |
136,820 - 137,570 (calc based on 18 Y, 20 W, and 6 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB: |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE: |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
146 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
51 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
72 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
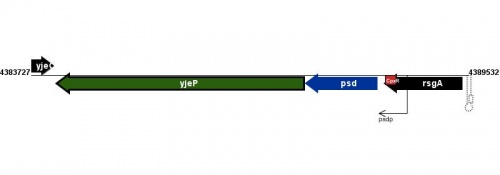 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:4387373..4387413
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for yjeP | |
|
microarray |
Summary of data for yjeP from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to yjeP Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
RegulonDB: |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP: |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Macaca mulatta |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
YJEP |
From SHIGELLACYC |
|
E. coli O157 |
YJEP |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc: |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene: |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB: |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE: |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP: |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
Categories
- TMHMM Prediction
- Genes with homologs in Apis mellifera
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Canis familiaris
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Gallus gallus
- Genes with homologs in Homo sapiens
- Genes with homologs in Macaca mulatta
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Oryza gramene
- Genes with homologs in Pan troglodytes
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157

