ptrA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
ptrA |
|---|---|
| Gene Synonym(s) | |
| Product Desc. |
Protease III, protease Pi, pitrilysin, zinc metalloprotease; perhaps some specificity for 10 to 30 residue peptides[4] |
| Product Synonyms(s) |
protease III[1], B2821[2][1], PtrA[2][1], Ptr[2][1], Pitrilysin[2][1], Protease pi[2][1] , ECK2817, JW2789, ptr, b2821 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
ptrA |
|---|---|
| Mnemonic |
Protease III |
| Synonyms | |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
63.67 minutes |
MG1655: 2956906..2954018 |
||
|
NC_012967: 2850615..2853503 |
||||
|
NC_012759: 2841166..2844054 |
||||
|
W3110 |
|
W3110: 2957540..2954652 |
||
|
DH10B: 3050776..3047888 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ptrAH88R |
H88R |
Loss of activity and of Zn-binding |
seeded from UniProt:P05458 | ||||
|
ptrAE162Q |
E162Q |
20% loss of activity |
seeded from UniProt:P05458 | ||||
|
ptrAE169Q |
E169Q |
Loss of activity and of Zn-binding |
seeded from UniProt:P05458 | ||||
|
ptrAE204Q |
E204Q |
No loss of activity |
seeded from UniProt:P05458 | ||||
|
ptrAE91Q |
E91Q |
Loss of activity |
seeded from UniProt:P05458 | ||||
|
ptrAH92R |
H92R |
Loss of activity and of Zn-binding |
seeded from UniProt:P05458 | ||||
|
Δptr::kan |
deletion |
Biolog:respiration |
unable to respire Acetate |
PMID:16095938 |
|||
|
Δptr::kan |
deletion |
Biolog:respiration |
unable to respire Bromosuccinate |
PMID:16095938 |
|||
|
Δptr::kan |
deletion |
Biolog:respiration |
unable to respire L-Asparagine |
PMID:16095938 |
|||
|
Δptr::kan |
deletion |
Biolog:respiration |
unable to respire L-Aspartate |
PMID:16095938 |
|||
|
ΔptrA746::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
Plasmid clone |
PMID:16769691 Status: Primer 1: Primer 2: | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10786 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10786 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000777 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0779 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0009252 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
Ptr |
|---|---|
| Synonyms |
protease III[1], B2821[2][1], PtrA[2][1], Ptr[2][1], Pitrilysin[2][1], Protease pi[2][1] , ECK2817, JW2789, ptr, b2821 |
| Product description |
Protease III, protease Pi, pitrilysin, zinc metalloprotease; perhaps some specificity for 10 to 30 residue peptides[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0030288 |
outer membrane-bounded periplasmic space |
C |
Seeded from Riley et al 2006 [1]. |
Missing: evidence, reference | ||||
|
GO:0000287 |
magnesium ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0460 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003824 |
catalytic activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011237 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0003824 |
catalytic activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011249 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004222 |
metalloendopeptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001431 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004222 |
metalloendopeptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007863 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0004222 |
metalloendopeptidase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011765 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006508 |
proteolysis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001431 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006508 |
proteolysis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007863 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006508 |
proteolysis |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011765 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008233 |
peptidase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0645 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008237 |
metallopeptidase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0482 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008270 |
zinc ion binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR007863 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008270 |
zinc ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0862 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016787 |
hydrolase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0378 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0042597 |
periplasmic space |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0574 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0042597 |
periplasmic space |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0200 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046872 |
metal ion binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011237 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046872 |
metal ion binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011249 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046872 |
metal ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0479 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
PMID:374413 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0006508 |
proteolysis |
PMID:374413 |
IDA: Inferred from Direct Assay |
P |
complete | |||
|
GO:0004222 |
metalloendopeptidase activity |
PMID:374413 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0030288 |
outer membrane-bounded periplasmic space |
PMID:7037737 |
IDA: Inferred from Direct Assay |
C |
complete | |||
|
GO:0008270 |
zinc ion binding |
PMID:1733942 |
IDA: Inferred from Direct Assay |
F |
complete | |||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
rplW |
PMID:16606699 |
Experiment(s):EBI-1144067 | |
|
Protein |
acpS |
PMID:16606699 |
Experiment(s):EBI-1144067 | |
|
Protein |
nadE |
PMID:16606699 |
Experiment(s):EBI-1144067 | |
|
Protein |
rplB |
PMID:16606699 |
Experiment(s):EBI-1144067 | |
|
Protein |
rplS |
PMID:16606699 |
Experiment(s):EBI-1144067 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
Periplasm |
PMID:15502359 |
| ||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MPRSTWFKAL LLLVALWAPL SQAETGWQPI QETIRKSDKD NRQYQAIRLD NGMVVLLVSD PQAVKSLSAL VVPVGSLEDP EAYQGLAHYL EHMSLMGSKK YPQADSLAEY LKMHGGSHNA STAPYRTAFY LEVENDALPG AVDRLADAIA EPLLDKKYAE RERNAVNAEL TMARTRDGMR MAQVSAETIN PAHPGSKFSG GNLETLSDKP GNPVQQALKD FHEKYYSANL MKAVIYSNKP LPELAKMAAD TFGRVPNKES KKPEITVPVV TDAQKGIIIH YVPALPRKVL RVEFRIDNNS AKFRSKTDEL ITYLIGNRSP GTLSDWLQKQ GLVEGISANS DPIVNGNSGV LAISASLTDK GLANRDQVVA AIFSYLNLLR EKGIDKQYFD ELANVLDIDF RYPSITRDMD YVEWLADTMI RVPVEHTLDA VNIADRYDAK AVKERLAMMT PQNARIWYIS PKEPHNKTAY FVDAPYQVDK ISAQTFADWQ KKAADIALSL PELNPYIPDD FSLIKSEKKY DHPELIVDES NLRVVYAPSR YFASEPKADV SLILRNPKAM DSARNQVMFA LNDYLAGLAL DQLSNQASVG GISFSTNANN GLMVNANGYT QRLPQLFQAL LEGYFSYTAT EDQLEQAKSW YNQMMDSAEK GKAFEQAIMP AQMLSQVPYF SRDERRKILP SITLKEVLAY RDALKSGARP EFMVIGNMTE AQATTLARDV QKQLGADGSE WCRNKDVVVD KKQSVIFEKA GNSTDSALAA VFVPTGYDEY TSSAYSSLLG QIVQPWFYNQ LRTEEQLGYA VFAFPMSVGR QWGMGFLLQS NDKQPSFLWE RYKAFFPTAE AKLRAMKPDE FAQIQQAVIT QMLQAPQTLG EEASKLSKDF DRGNMRFDSR DKIVAQIKLL TPQKLADFFH QAVVEPQGMA ILSQISGSQN GKAEYVHPEG WKVWENVSAL QQTMPLMSEK NE |
| Length |
962 |
| Mol. Wt |
107.711 kDa |
| pI |
6.0 (calculated) |
| Extinction coefficient |
135,110 - 135,235 (calc based on 39 Y, 14 W, and 1 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0009252 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10786 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10786 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000777 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0779 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
227 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
140 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
229 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
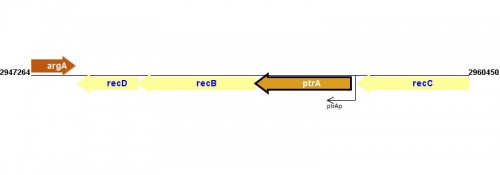 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:2956886..2956926
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for ptrA | |
|
microarray |
Summary of data for ptrA from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to ptrA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10786 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0779 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10786 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000777 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0009252 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Canis familiaris |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Drosophila melanogaster |
|
From Inparanoid:20070104 |
|
Drosophila pseudoobscura |
|
From Inparanoid:20070104 |
|
Gallus gallus |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Macaca mulatta |
|
From Inparanoid:20070104 |
|
Monodelphis domestica |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Pan troglodytes |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Saccharomyces cerevisiae |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Takifugu rubripes |
|
From Inparanoid:20070104 |
|
Tetraodon nigroviridis |
|
From Inparanoid:20070104 |
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
PTR |
From SHIGELLACYC |
|
E. coli O157 |
PTR |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10786 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10786 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000777 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0779 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0009252 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
Categories
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Apis mellifera
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Bos taurus
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Canis familiaris
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Drosophila melanogaster
- Genes with homologs in Drosophila pseudoobscura
- Genes with homologs in Gallus gallus
- Genes with homologs in Homo sapiens
- Genes with homologs in Macaca mulatta
- Genes with homologs in Monodelphis domestica
- Genes with homologs in Mus musculus
- Genes with homologs in Oryza gramene
- Genes with homologs in Pan troglodytes
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Saccharomyces cerevisiae
- Genes with homologs in Schizosaccharomyces pombe
- Genes with homologs in Takifugu rubripes
- Genes with homologs in Tetraodon nigroviridis
- Genes with homologs in Xenopus tropicalis
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


