mrcB:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
mrcB |
|---|---|
| Gene Synonym(s) |
ECK0148, b0149, JW0145, pbpF, ponB[1], ponB |
| Product Desc. |
Component of penicillin-binding protein 1B[3] Murein polymerase, PBP1B; bifunctional murein transglycosylase and transpeptidase; penicillin-binding protein 1B; dimeric[4] |
| Product Synonyms(s) |
fused glycosyl transferase[1], transpeptidase[1], B0149[2][1], PonB[2][1], PbpF[2][1], MrcB[2][1], PBP1B[2][1] , ECK0148, JW0145, pbpF, ponB, b0149 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
MrcB is part of a complex with MltA and MipA. The N-terminal transglycosylase activity polymerizes the glycan backbone and is penicillin-insensitive. The C-terminal transpeptidase activity crosslinks the muramic peptides and is penicillin-sensitive. Full length MrcB is the alpha isoform. A second in vivo ATG start at codon 46 produces the 799 aa gamma isoform. Murein polymerase is an essential function satisfied by either MrcA or MrcB. Mutants have elevated OM vesiculation (McBroom, 2006). MrcB is a membrane-bound periplasmic enzyme that has an uncleaved signal anchor.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
mrcB |
|---|---|
| Mnemonic |
Murein cluster c |
| Synonyms |
ECK0148, b0149, JW0145, pbpF, ponB[1], ponB |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
3.55 minutes |
MG1655: 164730..167264 |
||
|
NC_012967: 167581..170106 |
||||
|
NC_012759: 164729..167263 |
||||
|
W3110 |
|
W3110: 164730..167264 |
||
|
DH10B: 138834..141368 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
mrcB(del) (Keio:JW0145) |
deletion |
deletion |
PMID:16738554 |
||||
|
mrcBE233Q |
E233Q |
Loss of wild-type glycan chain elongation activity. No complementation in strain defective in PBP-1b |
seeded from UniProt:P02919 | ||||
|
mrcBD234N |
D234N |
7-fold decrease in catalytic activity. No complementation in strain defective in PBP-1b |
seeded from UniProt:P02919 | ||||
|
mrcBE290Q |
E290Q |
11-fold decrease in catalytic activity. Shows complementation activity in strain defective in PBP-1b |
seeded from UniProt:P02919 | ||||
|
mrcB704 |
|||||||
|
ΔmrcB765::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW0145 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCGCCGGGAATGACCGCGAGCC Primer 2:CCATTACTACCAAACATATCCTT | |
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 56% [6] | ||
|
Linked marker |
est. P1 cotransduction: 2% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10605 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10605 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000598 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0600 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000516 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
MrcB |
|---|---|
| Synonyms |
fused glycosyl transferase[1], transpeptidase[1], B0149[2][1], PonB[2][1], PbpF[2][1], MrcB[2][1], PBP1B[2][1] , ECK0148, JW0145, pbpF, ponB, b0149 |
| Product description |
Component of penicillin-binding protein 1B[3] Murein polymerase, PBP1B; bifunctional murein transglycosylase and transpeptidase; penicillin-binding protein 1B; dimeric[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0005887 |
integral to plasma membrane |
C |
Seeded from Riley et al 2006 [1]. |
Missing: evidence, reference | ||||
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0997 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005886 |
plasma membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-1003 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008955 |
peptidoglycan glycosyltransferase activity |
PMID:6389538 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0051518 |
positive regulation of bipolar cell growth |
PMID:330236 |
IDA: Inferred from Direct Assay |
P |
Cephaloridine has a high affinity for PBP1B. When this happens, cell elongation is inhibited, causing the cell to lyse. |
complete | ||
|
GO:0009002 |
serine-type D-Ala-D-Ala carboxypeptidase activity |
PMID:6389538 |
IDA: Inferred from Direct Assay |
F |
complete | |||
|
GO:0008658 |
penicillin binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001460 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008955 |
peptidoglycan glycosyltransferase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011813 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008955 |
peptidoglycan glycosyltransferase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:2.4.1.129 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009252 |
peptidoglycan biosynthetic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001264 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009252 |
peptidoglycan biosynthetic process |
PMID:16569861 |
IMP: Inferred from Mutant Phenotype |
P |
Table 1 shows a PBP1B deficient strain produce lipid II instead of peptidoglycan. |
complete | ||
|
GO:0009252 |
peptidoglycan biosynthetic process |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0573 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009274 |
peptidoglycan-based cell wall |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR001264 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009274 |
peptidoglycan-based cell wall |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011813 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0472 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
PMID:16858726 |
IDA: Inferred from Direct Assay |
C |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0016021 |
integral to membrane |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0812 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016021 |
integral to membrane |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-9906 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046677 |
response to antibiotic |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR011813 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046677 |
response to antibiotic |
PMID:371539 |
IDA: Inferred from Direct Assay |
SP_KW:KW-0046
|
P |
Table 1 shows Cephalosporin Beta-Lactams exhibit high affinity for PBP1B. |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of penicillin-binding protein 1B |
could be indirect |
||
|
Protein |
talB |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
hupA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
hemB |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
ppsA |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
plasma membrane |
| |||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MAGNDREPIG RKGKPTRPVK QKVSRRRYED DDDYDDYDDY EDEEPMPRKG KGKGKGRKPR GKRGWLWLLL KLAIVFAVLI AIYGVYLDQK IRSRIDGKVW QLPAAVYGRM VNLEPDMTIS KNEMVKLLEA TQYRQVSKMT RPGEFTVQAN SIEMIRRPFD FPDSKEGQVR ARLTFDGDHL ATIVNMENNR QFGFFRLDPR LITMISSPNG EQRLFVPRSG FPDLLVDTLL ATEDRHFYEH DGISLYSIGR AVLANLTAGR TVQGASTLTQ QLVKNLFLSS ERSYWRKANE AYMALIMDAR YSKDRILELY MNEVYLGQSG DNEIRGFPLA SLYYFGRPVE ELSLDQQALL VGMVKGASIY NPWRNPKLAL ERRNLVLRLL QQQQIIDQEL YDMLSARPLG VQPRGGVISP QPAFMQLVRQ ELQAKLGDKV KDLSGVKIFT TFDSVAQDAA EKAAVEGIPA LKKQRKLSDL ETAIVVVDRF SGEVRAMVGG SEPQFAGYNR AMQARRSIGS LAKPATYLTA LSQPKIYRLN TWIADAPIAL RQPNGQVWSP QNDDRRYSES GRVMLVDALT RSMNVPTVNL GMALGLPAVT ETWIKLGVPK DQLHPVPAML LGALNLTPIE VAQAFQTIAS GGNRAPLSAL RSVIAEDGKV LYQSFPQAER AVPAQAAYLT LWTMQQVVQR GTGRQLGAKY PNLHLAGKTG TTNNNVDTWF AGIDGSTVTI TWVGRDNNQP TKLYGASGAM SIYQRYLANQ TPTPLNLVPP EDIADMGVDY DGNFVCSGGM RILPVWTSDP QSLCQQSEMQ QQPSGNPFDQ SSQPQQQPQQ QPAQQEQKDS DGVAGWIKDM FGSN |
| Length |
844 |
| Mol. Wt |
94.294 kDa |
| pI |
9.5 (calculated) |
| Extinction coefficient |
116,200 - 116,450 (calc based on 30 Y, 13 W, and 2 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0000516 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10605 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10605 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000598 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0600 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
Ecoli K-12 |
0.945+/-0.065 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.023875296 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
521 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
139 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
259 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
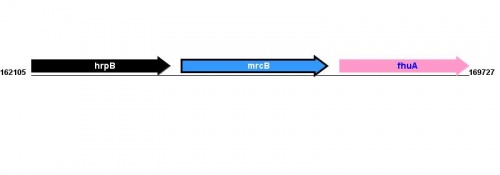 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:164710..164750
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for mrcB | |
|
microarray |
Summary of data for mrcB from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (164452..164755) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to mrcB Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10605 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0600 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10605 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000598 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000516 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Shigella flexneri |
MRCB |
From SHIGELLACYC |
|
E. coli O157 |
MRCB |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10605 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10605 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000598 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0600 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0000516 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories


