gltB:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
gltB |
|---|---|
| Gene Synonym(s) |
ECK3202, b3212, JW3179, aspB, ossB, psiQ[1], psiQ |
| Product Desc. |
glutamate synthase (NADPH) large chain precursor[2]; glutamate synthase, large subunit[3]; Component of glutamate synthase[2][3]; glutamate synthase (NADPH)[2] Glutamate synthase, large subunit[4] |
| Product Synonyms(s) |
glutamate synthase, large subunit[1], B3212[2][1], OssB[2][1], PsiQ[2][1], AspB[2][1], GltB[2][1] , aspB, ECK3202, JW3179, ossB, psiQ, b3212 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
gltB |
|---|---|
| Mnemonic |
Glutamate |
| Synonyms |
ECK3202, b3212, JW3179, aspB, ossB, psiQ[1], psiQ |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
72.26 minutes |
MG1655: 3352654..3357207 |
||
|
NC_012967: 3289867..3294420 |
||||
|
NC_012759: 3239802..3244355 |
||||
|
W3110 |
|
W3110: 3354487..3359040 |
||
|
DH10B: 3450399..3454952 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔgltB (Keio:JW3179) |
deletion |
deletion |
PMID:16738554 |
||||
|
gltB17 |
PMID:374339 |
||||||
|
gltB13 |
|||||||
|
gltB32 |
|||||||
|
ΔgltB740::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW3179 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCACACGCAAACCCCGTCGCCA Primer 2:CCtTGCGCCTGCACGCGCAACTC | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 46% [6] | ||
|
Linked marker |
est. P1 cotransduction: 68% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10403 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10403 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000396 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB0398 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010545 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
GltB |
|---|---|
| Synonyms |
glutamate synthase, large subunit[1], B3212[2][1], OssB[2][1], PsiQ[2][1], AspB[2][1], GltB[2][1] , aspB, ECK3202, JW3179, ossB, psiQ, b3212 |
| Product description |
glutamate synthase (NADPH) large chain precursor[2]; glutamate synthase, large subunit[3]; Component of glutamate synthase[2][3]; glutamate synthase (NADPH)[2] Glutamate synthase, large subunit[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0005737 |
cytoplasm |
C |
Seeded from Riley et al 2006 [1]. |
Missing: evidence, reference | ||||
|
GO:0004355 |
glutamate synthase (NADPH) activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:1.4.1.13 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005506 |
iron ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0408 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006537 |
glutamate biosynthetic process |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002932 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006537 |
glutamate biosynthetic process |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0314 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006541 |
glutamine metabolic process |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0315 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046872 |
metal ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0479 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0051536 |
iron-sulfur cluster binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0411 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0051538 |
3 iron, 4 sulfur cluster binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0003 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR002489 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006982 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0560 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of glutamate synthase |
could be indirect |
||
|
Protein |
katG |
PMID:19402753 |
LCMS(ID Probability):99.6 | |
|
Protein |
glpK |
PMID:19402753 |
MALDI(Z-score):24.217330 | |
|
Protein |
ebgA |
PMID:19402753 |
MALDI(Z-score):17.480125 | |
|
Protein |
gltD |
PMID:19402753 |
LCMS(ID Probability):99.6 MALDI(Z-score):25.464654 | |
|
Protein |
Subunits of glutamate synthase (NADPH) |
could be indirect |
| |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MTRKPRRHAL SVPVRSGSEV GFPQSLGEVH DMLYDKSLER DNCGFGLIAH IEGEPSHKVV RTAIHALARM QHRGAILADG KTGDGCGLLL QKPDRFFRIV AQERGWRLAK NYAVGMLFLN KDPELAAAAR RIVEEELQRE TLSIVGWRDV PTNEGVLGEI ALSSLPRIEQ IFVNAPAGWR PRDMERRLFI ARRRIEKRLE ADKDFYVCSL SNLVNIYKGL CMPTDLPRFY LDLADLRLES AICLFHQRFS TNTVPRWPLA QPFRYLAHNG EINTITGNRQ WARARTYKFQ TPLIPDLHDA APFVNETGSD SSSMDNMLEL LLAGGMDIIR AMRLLVPPAW QNNPDMDPEL RAFFDFNSMH MEPWDGPAGI VMSDGRFAAC NLDRNGLRPA RYVITKDKLI TCASEVGIWD YQPDEVVEKG RVGPGELMVI DTRSGRILHS AETDDDLKSR HPYKEWMEKN VRRLVPFEDL PDEEVGSREL DDDTLASYQK QFNYSAEELD SVIRVLGENG QEAVGSMGDD TPFAVLSSQP RIIYDYFRQQ FAQVTNPPID PLREAHVMSL ATSIGREMNV FCEAEGQAHR LSFKSPILLY SDFKQLTTMK EEHYRADTLD ITFDVTKTTL EATVKELCDK AEKMVRSGTV LLVLSDRNIA KDRLPVPAPM AVGAIQTRLV DQSLRCDANI IVETASARDP HHFAVLLGFG ATAIYPYLAY ETLGRLVDTH AIAKDYRTVM LNYRNGINKG LYKIMSKMGI STIASYRCSK LFEAVGLHDD VVGLCFQGAV SRIGGASFED FQQDLLNLSK RAWLARKPIS QGGLLKYVHG GEYHAYNPDV VRTLQQAVQS GEYSDYQEYA KLVNERPATT LRDLLAITPG ENAVNIADVE PASELFKRFD TAAMSIGALS PEAHEALAEA MNSIGGNSNS GEGGEDPARY GTNKVSRIKQ VASGRFGVTP AYLVNADVIQ IKVAQGAKPG EGGQLPGDKV TPYIAKLRYS VPGVTLISPP PHHDIYSIED LAQLIFDLKQ VNPKAMISVK LVSEPGVGTI ATGVAKAYAD LITIAGYDGG TGASPLSSVK YAGCPWELGL VETQQALVAN GLRHKIRLQV DGGLKTGVDI IKAAILGAES FGFGTGPMVA LGCKYLRICH LNNCATGVAT QDDKLRKNHY HGLPFKVTNY FEFIARETRE LMAQLGVTRL VDLIGRTDLL KELDGFTAKQ QKLALSKLLE TAEPHPGKAL YCTENNPPFD NGLLNAQLLQ QAKPFVDERQ SKTFWFDIRN TDRSVGASLS GYIAQTHGDQ GLAADPIKAY FNGTAGQSFG VWNAGGVELY LTGDANDYVG KGMAGGLIAI RPPVGSAFRS HEASIIGNTC LYGATGGRLY AAGRAGERFG VRNSGAITVV EGIGDNGCEY MTGGIVCILG KTGVNFGAGM TGGFAYVLDE SGDFRKRVNP ELVEVLSVDA LAIHEEHLRG LITEHVQHTG SQRGEEILAN WSTFATKFAL VKPKSSDVKA LLGHRSRSAA ELRVQAQ |
| Length |
1,517 |
| Mol. Wt |
166.713 kDa |
| pI |
6.7 (calculated) |
| Extinction coefficient |
150,010 - 152,510 (calc based on 49 Y, 14 W, and 20 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0010545 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG10403 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10403 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120000396 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0398 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
7.29E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
Ecoli K-12 |
174.805+/-0.994 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.10845+/-0.01198 |
Molecules/cell |
|
by FISH |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.362398419 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
1079 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
6013 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
1004 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
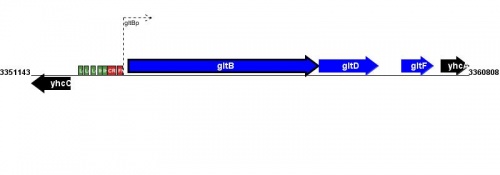 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:3352634..3352674
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for gltB | |
|
microarray |
Summary of data for gltB from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (3351589..3352343) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to gltB Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10403 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0398 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10403 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000396 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010545 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Apis mellifera |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Caenorhabditis briggsae |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Drosophila melanogaster |
|
From Inparanoid:20070104 |
|
Drosophila pseudoobscura |
|
From Inparanoid:20070104 |
|
Oryza gramene |
|
From Inparanoid:20070104 |
|
Saccharomyces cerevisiae |
|
From Inparanoid:20070104 |
|
Schizosaccharomyces pombe |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
GLTB |
From SHIGELLACYC |
|
E. coli O157 |
GLTB |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG10403 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG10403 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120000396 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB0398 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0010545 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories
- Genes in OpenBioSystems with Promoter Fusions
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Apis mellifera
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Caenorhabditis briggsae
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Drosophila melanogaster
- Genes with homologs in Drosophila pseudoobscura
- Genes with homologs in Oryza gramene
- Genes with homologs in Saccharomyces cerevisiae
- Genes with homologs in Schizosaccharomyces pombe
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157


