fdoG:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
fdoG |
|---|---|
| Gene Synonym(s) |
ECK3887, b3894, JW3865[1], JW3865 |
| Product Desc. |
formate dehydrogenase-O, α subunit[2][3]; Component of formate dehydrogenase-O[2][3] Formate dehydrogenase-O, selenopeptide, aerobic[4] |
| Product Synonyms(s) |
formate dehydrogenase-O, large subunit[1], B3894[2][1], FdoG[2][1] , ECK3887, JW3865, b3894 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression |
transcription unit(s): fdoGHI-fdhE[2] |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Also known as FDH-Z. Verified Tat substrate: FdoG has a predicted Tat-only (Class I) 33 aa signal peptide (Tullman-Ercek, 2007). Binds TrxA (Kumar, 2004). HT_Cmplx13_Mem: FdoG+FdoH. The UGA stop codon 196 is translated as selenocysteine in vivo and indicated as a "U" in the protein sequence.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
fdoG |
|---|---|
| Mnemonic |
Formate dehydrogenase-O |
| Synonyms |
ECK3887, b3894, JW3865[1], JW3865 |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
87.95 minutes |
MG1655: 4083845..4080795 |
||
|
NC_012967: 4063969..4060919 |
||||
|
NC_012759: 3970464..3973514 |
||||
|
W3110 |
|
W3110: 3550859..3553909 |
||
|
DH10B: 4183542..4180492 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
Strain DH10B |
N307T |
Nonsynonomous mutation |
PMID:18245285 |
This is a SNP in E. coli K-12 Strain DH10B | |||
|
ΔfdoG (Keio:JW3865) |
deletion |
deletion |
PMID:16738554 |
||||
|
ΔfdoG757::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
JW3865 |
Plasmid clone |
PMID:16769691 Status:Clone OK Primer 1:GCCCAGGTCAGCAGAAGGCAGTT Primer 2:CCaACCTTTTCCACATTCACAAG | |
|
Kohara Phage |
PMID:3038334 | ||
|
Kohara Phage |
PMID:3038334 | ||
|
Linked marker |
est. P1 cotransduction: 56% [6] | ||
|
Linked marker |
est. P1 cotransduction: 4% [6] | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11858 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11858 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001789 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB1804 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012710 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
FdoG |
|---|---|
| Synonyms |
formate dehydrogenase-O, large subunit[1], B3894[2][1], FdoG[2][1] , ECK3887, JW3865, b3894 |
| Product description |
formate dehydrogenase-O, α subunit[2][3]; Component of formate dehydrogenase-O[2][3] Formate dehydrogenase-O, selenopeptide, aerobic[4] |
| EC number (for enzymes) | |
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0030288 |
outer membrane-bounded periplasmic space |
C |
Seeded from Riley et al 2006 [1]. |
Missing: evidence, reference | ||||
|
GO:0005506 |
iron ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0408 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0005737 |
cytoplasm |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006443 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008430 |
selenium binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0711 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008863 |
formate dehydrogenase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006443 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0008863 |
formate dehydrogenase activity |
GOA:spec |
IEA: Inferred from Electronic Annotation |
EC:1.2.1.2 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0009055 |
electron carrier activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006655 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016020 |
membrane |
PMID:16858726 |
IDA: Inferred from Direct Assay |
C |
Seeded from EcoCyc (v14.0) |
complete | ||
|
GO:0016491 |
oxidoreductase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006655 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016491 |
oxidoreductase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006656 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016491 |
oxidoreductase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006657 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016491 |
oxidoreductase activity |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006963 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0016491 |
oxidoreductase activity |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0560 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030151 |
molybdenum ion binding |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006657 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0030151 |
molybdenum ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0500 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0042597 |
periplasmic space |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0574 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0042597 |
periplasmic space |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0200 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0045333 |
cellular respiration |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006443 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0046872 |
metal ion binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0479 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0051536 |
iron-sulfur cluster binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0411 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0051539 |
4 iron, 4 sulfur cluster binding |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0004 |
F |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:interpro |
IEA: Inferred from Electronic Annotation |
InterPro:IPR006443 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0055114 |
oxidation reduction |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0560 |
P |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0006974 |
response to DNA damage stimulus |
PMID:11967071 |
IEP: Inferred from Expression Pattern |
P |
complete | |||
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
Subunits of formate dehydrogenase-O |
could be indirect |
||
|
Protein |
nadE |
PMID:16606699 |
Experiment(s):EBI-1146913 | |
|
Protein |
polA |
PMID:16606699 |
Experiment(s):EBI-1146913 | |
|
Protein |
ileS |
PMID:16606699 |
Experiment(s):EBI-1146913 | |
|
Protein |
cyaA |
PMID:16606699 |
Experiment(s):EBI-1146913 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
|
Periplasm |
PMID:12923080 |
|||
|
cytoplasm |
From EcoCyc[3] |
| ||
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MQVSRRQFFK ICAGGMAGTT AAALGFAPSV ALAETRQYKL LRTRETRNTC TYCSVGCGLL MYSLGDGAKN AKASIFHIEG DPDHPVNRGA LCPKGAGLVD FIHSESRLKF PEYRAPGSDK WQQISWEEAF DRIAKLMKED RDANYIAQNA EGVTVNRWLS TGMLCASASS NETGYLTQKF SRALGMLAVD NQARVUHGPT VASLAPTFGR GAMTNHWVDI KNANLVVVMG GNAAEAHPVG FRWAMEAKIH NGAKLIVIDP RFTRTAAVAD YYAPIRSGTD IAFLSGVLLY LLNNEKFNRE YTEAYTNASL IVREDYGFED GLFTGYDAEK RKYDKSSWTY ELDENGFAKR DTTLQHPRCV WNLLKQHVSR YTPDVVENIC GTPKDAFLKV CEYIAETSAH DKTASFLYAL GWTQHSVGAQ NIRTMAMIQL LLGNMGMAGG GVNALRGHSN IQGLTDLGLL SQSLPGYMTL PSEKQTDLQT YLTANTPKPL LEGQVNYWGN YPKFFVSMMK AFFGDKATAE NSWGFDWLPK WDKGYDVLQY FEMMKEGKVN GYICQGFNPV ASFPNKNKVI GCLSKLKFLV TIDPLNTETS NFWQNHGELN EVDSSKIQTE VFRLPSTCFA EENGSIVNSG RWLQWHWKGA DAPGIALTDG EILSGIFLRL RKMYAEQGGA NPDQVLNMTW NYAIPHEPSS EEVAMESNGK ALADITDPAT GAVIVKKGQQ LSSFAQLRDD GTTSCGCWIF AGSWTPEGNQ MARRDNADPS GLGNTLGWAW AWPLNRRILY NRASADPQGN PWDPKRQLLK WDGTKWTGWD IPDYSAAPPG SGVGPFIMQQ EGMGRLFALD KMAEGPFPEH YEPFETPLGT NPLHPNVISN PAARIFKDDA EALGKADKFP YVGTTYRLTE HFHYWTKHAL LNAILQPEQF VEIGESLANK LGIAQGDTVK VSSNRGYIKA KAVVTKRIRT LKANGKDIDT IGIPIHWGYE GVAKKGFIAN TLTPFVGDAN TQTPEFKSFL VNVEKV |
| Length |
1,015 |
| Mol. Wt |
112.401 kDa |
| pI |
7.3 (calculated) |
| Extinction coefficient |
206,150 - 207,900 (calc based on 35 Y, 28 W, and 14 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0012710 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG11858 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11858 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120001789 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1804 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MC4100 |
6.72E+02 |
molecules/cell |
|
emPAI |
PMID:18304323 | |
|
Protein |
E. coli K-12 MG1655 |
219 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
11 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
37 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
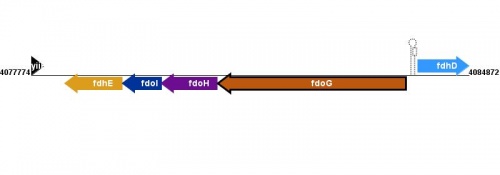 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:4083825..4083865
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for fdoG | |
|
microarray |
Summary of data for fdoG from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
|
GFP Fusion |
Intergenic region (4083317..4083666) fused to gfpmut2. |
GFP fusion described in Zaslaver, et al. | ||
| edit table |
<protect></protect>
Notes
Accessions Related to fdoG Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11858 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1804 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11858 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001789 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012710 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Xenopus tropicalis |
|
From Inparanoid:20070104 |
|
E. coli O157 |
FDOG |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG11858 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG11858 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120001789 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB1804 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0012710 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 3.2 3.3 3.4 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 5.2 CGSC: The Coli Genetics Stock Center
- ↑ 6.0 6.1 The Tn10 insertion sites determined by Nichols et al. 1998 (PMID:9829956) were reannotated by alignment with E. coli K-12 genome sequence (GenBank accession NC_000913). P1 contransduction frequencies were calculated using the formula of Wu (PMID:5338813).
- ↑ Zaslaver, A et al. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3 623-8 PubMed
Categories


