ychO:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
ychO |
|---|---|
| Gene Synonym(s) | |
| Product Desc. |
Intimin/invasin homolog, function unknown[4] |
| Product Synonyms(s) |
predicted invasin[1], B1220[2][1], YchO[2][1], YchP[2][1] , ECK1214, JW1211, ychP, b1220 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Extension based on homology to S. typhimurium SinH. YchO has a 24 aa signal peptide predicted.[4]
Gene
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
Nomenclature
See Help:Gene_nomenclature for help with entering information in the Gene Nomenclature table.
| Standard name |
ychO |
|---|---|
| Mnemonic |
Systematic nomenclature |
| Synonyms | |
| edit table |
</protect>
Notes
Location(s) and DNA Sequence
<protect>
See Help:Gene_location for help entering information in the Gene Location and DNA sequence table.
| Strain | Map location | Genome coordinates | Genome browsers | Sequence links |
|---|---|---|---|---|
|
MG1655 |
27.44 minutes |
MG1655: 1273148..1274401 |
||
|
NC_012967: 1274075..1275328 |
||||
|
NC_012759: 1161728..1163122 |
||||
|
W3110 |
|
W3110: 1275502..1276755 |
||
|
DH10B: 1313248..1314642 |
||||
| edit table |
</protect>
Notes
Sequence Features
See Help:Gene_sequence_features for help in entering sequence features in EcoliWiki.
| Feature Type | Strain | Genomic Location | Evidence | Reference | Notes |
|---|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Alleles and Phenotypes
See Help:Gene_alleles for how to enter or edit alleles and phenotypes in EcoliWiki.
| Allele | Nt change(s) | AA change(s) | Phenotype: Type | Phenotype: Description | Reference | Availability | Comments |
|---|---|---|---|---|---|---|---|
|
ΔychO731::kan |
PMID:16738554 |
| |||||
| edit table |
<protect></protect>
Notes
Phenotypes of Escherichia coli APEC SEPT362 (NCBI Taxonomy ID:1196033) ychO(del) described in Pilatti, L. et al. (2016).[5]
- reduced adhesion to chicken embryo fibroblast cells in both the presence and absence of methyl-alpha-D[mannopyranoside (Fig. 2a)
- Expression of genes important for adhesion (aatA, aatB, fimH, eco, csgA, and lpfA) was analyzed by qRT-PCR analysis (Fig 2b) No statistically significant differences in expression level were seen in the 3 strains analyzed: SEPT362, SEPT362 ychO(del), and SEPT36s ychO(del)-complemented with plasmid borne ychO+.
- reduced invasion of cells from the chick embryo fibroblast cell line CEC-32 (Fig 3)
- reduced biofilm formation (assayed in 24-well polystyrene plates and quantified by crystal violet staining) (Fig.4)
- the authors conclude that ychO is important for virulence of APEC SEPT362 (Fig. 5)
- at day 7 post-inoculation, no statistically significant difference was seen in survival of chicks inoculated with SEPT362, SEPT362 ychO(del), or SEPT362 ychO(del) p(ychO))
- survival of chicks inoculated with SEPT362 ychO(del) was higher at day 4 and day 6 of the assay
- transcriptome sequencing (RNAseq) of SEPT362 and SEPT362 ychO(del) identified 426 genes whose transcript levels were altered
- among the altered genes, 94% were down regulated in the mutant (Fig. 6b)
- affected genes were grouped by functional category (Fig. 6a) but the paper did not include a list of the 426 genes whose transcript levels were altered
- RNAseq results were validated by comparison of the expression levels of 9 genes (csgA, lpfA, fimH, clic, flhD, motA, flgE, eco, and ychO) by qRT-PCR and the results were similar for both methods (data not shown)
Genetic Interactions
<protect>
| Interactor | Interaction | Allele | Score(s) | Reference(s) | Accessions | Notes |
|---|---|---|---|---|---|---|
| edit table |
</protect>
Notes
Genetic Resources
See Help:Gene_resources for help entering information into the Genetic Resources table.
| Resource | Resource Type | Source | Notes/Reference |
|---|---|---|---|
|
Plasmid clone |
PMID:16769691 Status: Primer 1: Primer 2: | ||
|
Kohara Phage |
PMID:3038334 | ||
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG12405 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12405 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120002267 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EchoBASE:EB2305 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0004095 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
YchP |
|---|---|
| Synonyms |
predicted invasin[1], B1220[2][1], YchO[2][1], YchP[2][1] , ECK1214, JW1211, ychP, b1220 |
| Product description |
Intimin/invasin homolog, function unknown[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
|
GO:0042597 |
periplasmic space |
GOA:spkw |
IEA: Inferred from Electronic Annotation |
SP_KW:KW-0574 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
|
GO:0042597 |
periplasmic space |
GO_REF:0000023 |
IEA: Inferred from Electronic Annotation |
SP_SL:SL-0200 |
C |
Seeded from EcoCyc (v14.0) |
complete | |
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
rhsA |
PMID:16606699 |
Experiment(s):EBI-1139210 | |
|
Protein |
groL |
PMID:16606699 |
Experiment(s):EBI-1139210 | |
|
Protein |
rhsC |
PMID:16606699 |
Experiment(s):EBI-1139210 | |
|
Protein |
nadE |
PMID:16606699 |
Experiment(s):EBI-1139210 | |
|
Protein |
mreB |
PMID:16606699 |
Experiment(s):EBI-1139210 | |
|
Protein |
rsuA |
PMID:16606699 |
Experiment(s):EBI-1139210 | |
|
Protein |
aspS |
PMID:16606699 |
Experiment(s):EBI-1139210 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MAPENHDGEK HFAEIVKDFG ETSMNDNGLD TGEQAKAFAL GKVRDALSQQ VNQHVESWLS PWGNASVDVK VDNEGHFTGS RGSWFVPLQD NDRYLTWSQL GLTQQDNGLV SNVGVGQRWA RGNWLVGYNT FYDNLLDENL QRAGFGAEAW GEYLRLSANF YQPFAAWHEQ TATQEQRMAR GYDLTARMRM PFYQHLNTSV SLEQYFGDRV DLFNSGTGYH NPVALSLGLN YTPVPLVTVT AQHKQGESGE NQNNLGLNLN YRFGVPLKKQ LSAGEVAESQ SLRGSRYDNP QRNNLPTLEY RQRKTLTVFL ATPPWDLKPG ETVPLKLQIR SRYGIRQLIW QGDTQILSLT PGAQANSAEG WTLIMPDWQN GEGASNHWRL SVVVEDNQGQ RVSSNEITLT LVEPFDALSN DELRWEP |
| Length |
417 |
| Mol. Wt |
47.022 kDa |
| pI |
5.1 (calculated) |
| Extinction coefficient |
97,860 (calc based on 14 Y, 14 W, and 0 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
ASAP:ABE-0004095 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
EcoCyc:EG12405 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12405 |
Escherichia coli str. K-12 substr. MG1655 | |
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB:ECK120002267 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2305 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
E. coli K-12 MG1655 |
26 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
21 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
12a |
molecules/cell/generation |
|
Ribosome Profiling |
Low confidence in the sequencing data set. |
PMID: 24766808 |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
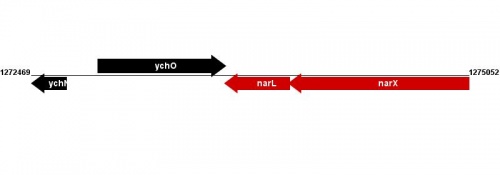 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
ychO in APEC strain SEPT362 (NCBI Taxonomy ID:1196033) is expressed in culture in LB medium and is also expressed in the spleen and lungs of chicks 24 and 48 h post-inoculation.[5]
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:1273128..1273168
source=MG1655
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for ychO | |
|
microarray |
Summary of data for ychO from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to ychO Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc:EG12405 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE:EB2305 |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene:EG12405 |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB:ECK120002267 |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP:ABE-0004095 |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Shigella flexneri |
YCHP |
From SHIGELLACYC |
|
E. coli O157 |
YCHP |
From ECOO157CYC |
| edit table |
<protect></protect>
Notes
Families
<protect>
See Help:Evolution_families for help entering or editing information in this section of EcoliWiki.
| Database | Accession | Notes |
|---|---|---|
|
| ||
| edit table |
</protect> <protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 4.2 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
- ↑ 5.0 5.1 Pilatti, L et al. (2016) The virulence factor ychO has a pleiotropic action in an Avian Pathogenic Escherichia coli (APEC) strain. BMC Microbiol. 16 35 PubMed
Categories


