tmcA:On One Page
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;} h2 .editsection { display:none;}</css>
| Quickview | Gene | Gene Product(s) | Expression | Evolution | On One Page |
| Quickview | | Gene | | Product(s) | | Expression| | Evolution | | References | |
Quickview
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
<protect>
| Standard Name |
ypfI |
|---|---|
| Gene Synonym(s) |
ECK2470, b2474, JW2459[1] |
| Product Desc. |
Function unknown; contains putative acetyltransferase domain[4] |
| Product Synonyms(s) |
predicted hydrolase[1], B2474[2][1], TmcA[2][1] , ECK2470, JW2459, ypfI, b2474 |
| Function from GO |
<GO_nr /> |
| Knock-Out Phenotype | |
| Regulation/Expression | |
| Regulation/Activity | |
| Quick Links | |
| edit table |
</protect> See Help:Quickview for help with entering information in the Quickview table. <protect></protect>
Notes
Gene
The database did not find the text of a page that it should have found, named "tmcA:Gene" .
This is usually caused by following an outdated diff or history link to a page that has been deleted.
If this is not the case, you may have found a bug in the software. Please report this to an administrator, making note of the URL.
Product(s)
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Nomenclature
See Help:Product_nomenclature for help entering or editing information in this section of EcoliWiki.
| Standard name |
TmcA |
|---|---|
| Synonyms |
predicted hydrolase[1], B2474[2][1], TmcA[2][1] , ECK2470, JW2459, ypfI, b2474 |
| Product description |
Function unknown; contains putative acetyltransferase domain[4] |
| EC number (for enzymes) |
|
| edit table |
<protect></protect>
Notes
Function
<protect>
Gene Ontology
See Help:Gene_ontology for help entering or editing GO terms and GO annotations in EcoliWiki.
| Qualifier | GO ID | GO term name | Reference | Evidence Code | with/from | Aspect | Notes | Status |
|---|---|---|---|---|---|---|---|---|
| edit table |
Interactions See Help:Product_interactions for help entering or editing information about gene product interactions in this section of EcoliWiki.
| Partner Type | Partner | Notes | References | Evidence |
|---|---|---|---|---|
|
Protein |
ycbB |
PMID:16606699 |
Experiment(s):EBI-1142979 | |
|
Protein |
yeaY |
PMID:16606699 |
Experiment(s):EBI-1142979 | |
|
Protein |
groL |
PMID:16606699 |
Experiment(s):EBI-1142979 | |
|
Protein |
ydfQ |
PMID:16606699 |
Experiment(s):EBI-1142979 | |
|
Protein |
dnaE |
PMID:16606699 |
Experiment(s):EBI-1142979 | |
|
Protein |
moaC |
PMID:16606699 |
Experiment(s):EBI-1142979 | |
|
Protein |
rpsE |
PMID:16606699 |
Experiment(s):EBI-1142979 | |
|
Protein |
mqo |
PMID:16606699 |
Experiment(s):EBI-1142979 | |
|
Protein |
groS |
PMID:19402753 |
LCMS(ID Probability):97.9 MALDI(Z-score):14.514901 | |
| edit table |
</protect>
Notes
Localization
See Help:Product_localization for how to add or edit information in this section of EcoliWiki.
| Compartment | Description | Evidence | Reference/Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Structure and Physical Properties
<protect>
Physical Properties
See Help:Product_physical_properties for help entering or editing information about the physical properties of this gene product.
| Name | |
|---|---|
| Sequence |
MAELTALHTL TAQMKREGIR RLLVLSGEEG WCFEHTLKLR DALPGDWLWI SPRPDAENHC SPSALQTLLG REFRHAVFDA RHGFDAAAFA ALSGTLKAGS WLVLLLPVWE EWENQPDADS LRWSDCPDPI ATPHFVQHLK RVLTADNEAI LWRQNQPFSL AHFTPRTDWY PATGAPQPEQ QQLLKQLMTM PPGVAAVTAA RGRGKSALAG QLISRIAGRA IVTAPAKAST DVLAQFAGEK FRFIAPDALL ASDEQADWLV VDEAAAIPAP LLHQLVSRFP RTLLTTTVQG YEGTGRGFLL KFCARFPHLH RFELQQPIRW AQGCPLEKMV SEALVFDDEN FTHTPQGNIV ISAFEQTLWQ SDPETPLKVY QLLSGAHYRT SPLDLRRMMD APGQHFLQAA GENEIAGALW LVDEGGLSQQ LSQAVWAGFR RPRGNLVAQS LAAHGNNPLA ATLRGRRVSR IAVHPARQRE GTGRQLIAGA LQYTQDLDYL SVSFGYTGEL WRFWQRCGFV LVRMGNHREA SSGCYTAMAL LPMSDAGKQL AEREHYRLRR DAQALAQWNG ETLPVDPLND AVLSDDDWLE LAGFAFAHRP LLTSLGCLLR LLQTSELALP ALRGRLQKNA SDAQLCTTLK LSGRKMLLVR QREEAAQALF ALNDVRTERL RDRITQWQLF H |
| Length |
671 |
| Mol. Wt |
74.893 kDa |
| pI |
7.7 (calculated) |
| Extinction coefficient |
117,910 - 119,035 (calc based on 9 Y, 19 W, and 9 C residues) |
| edit table |
Domains/Motifs/Modification Sites
|
See Help:Product_domains_motifs for help entering or editing information in this section of EcoliWiki.
|
<motif_map/> |
|
Structure
| </protect>
Structure figures<protect> | ||||||
Notes
Gene Product Resources
See Help:Product_resources for help with entering or editing information in this section of EcoliWiki.
| Resource type | Source | Notes/Reference |
|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions in Other Databases
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
Escherichia coli str. K-12 substr. MG1655 | ||
|
RegulonDB: |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE: |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Expression
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Overview
This is a placeholder for a summary statement on how expression of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Cellular Levels
| Molecule | Organism or Strain | Value | Units | Experimental Conditions | Assay used | Notes | Reference(s) |
|---|---|---|---|---|---|---|---|
|
Protein |
Ecoli K-12 |
2.615+/-0.2 |
Molecules/cell |
|
Single Molecule Fluorescence |
PMID:20671182 | |
|
mRNA |
Ecoli K-12 |
0.0191067 |
Molecules/cell |
|
by RNA_Seq |
PMID:20671182 | |
|
Protein |
E. coli K-12 MG1655 |
53 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
29 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
|
Protein |
E. coli K-12 MG1655 |
41 |
molecules/cell/generation |
|
Ribosome Profiling |
PMID: 24766808 | |
| edit table |
Notes
Transcription and Transcriptional Regulation
<protect>
|
See Help:Expression_transcription for help entering or editing information in this section of EcoliWiki.
|
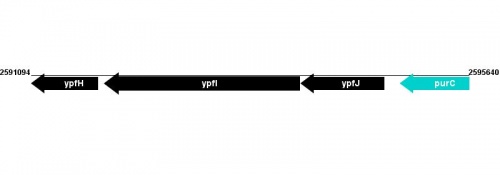 Figure courtesy of RegulonDB |
</protect>
Notes
This is a placeholder for a summary statement about how transcription of this gene is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Translation and Regulation of Translation
<protect><gbrowseImage>
name=NC_000913:2593861..2593901
source=MG1655
flip=1
type=Gene+DNA_+Protein
preset=Nterminus
</gbrowseImage>
This picture shows the sequence around the N-terminus.
</protect>
Notes
This is a placeholder for a summary statement about how translation of this gene product is regulated. You can help EcoliWiki by becoming a user and writing/editing this statement.
Turnover and Regulation of Turnover
</protect>
Notes
This is a placeholder for a summary statement about turnover of this gene product. You can help EcoliWiki by becoming a user and writing/editing this statement.
Experimental
<protect>
Mutations Affecting Expression
See Help:Expression_mutations for help entering or editing information in this section of EcoliWiki.
| Allele Name | Mutation | Phenotype | Reference |
|---|---|---|---|
| edit table |
Expression Studies
See Help:Expression_studies for help entering or editing information in this section of EcoliWiki.
| Type | Reference | Notes |
|---|---|---|
|
microarray |
NCBI GEO profiles for ypfI | |
|
microarray |
Summary of data for ypfI from multiple microarray studies | |
| edit table |
Expression Resources
See Help:Expression_resources for help entering or editing information in this section of EcoliWiki.
| Resource Name | Resource Type | Description | Source | Notes |
|---|---|---|---|---|
| edit table |
<protect></protect>
Notes
Accessions Related to tmcA Expression
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
RegulonDB: |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP: |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
Evolution
{{#css:Suppresslinks.css}}<css>h1 .editsection { display:none;}
h2 .editsection { display:none;}</css>
Homologs in Other Organisms
See Help:Evolution_homologs for help entering or editing information in this section of EcoliWiki.
| Organism | Homologs (Statistics) | Comments |
|---|---|---|
|
Anopheles gambiae |
|
From Inparanoid:20070104 |
|
Arabidopsis thaliana |
|
From Inparanoid:20070104 |
|
Bos taurus |
|
From Inparanoid:20070104 |
|
Caenorhabditis elegans |
|
From Inparanoid:20070104 |
|
Ciona intestinalis |
|
From Inparanoid:20070104 |
|
Dictyostelium discoideum |
|
From Inparanoid:20070104 |
|
Homo sapiens |
|
From Inparanoid:20070104 |
|
Mus musculus |
|
From Inparanoid:20070104 |
|
Rattus norvegicus |
|
From Inparanoid:20070104 |
|
Shigella flexneri |
YPFI |
From SHIGELLACYC |
|
E. coli O157 |
YPFI |
From ECOO157CYC |
| edit table |
Do-It-Yourself Web Tools
<protect></protect>
Notes
Families
See Help:Gene_accessions for help with entering information into the Gene Accessions table.
| Database | Accession | Notes |
|---|---|---|
|
EcoCyc: |
Escherichia coli str. K-12 substr. MG1655 | |
|
EcoGene: |
Escherichia coli str. K-12 substr. MG1655 | |
|
RegulonDB: |
Escherichia coli str. K-12 substr. MG1655 | |
|
EchoBASE: |
Escherichia coli str. K-12 substr. MG1655 | |
|
ASAP: |
Escherichia coli str. K-12 substr. MG1655 | |
| edit table |
<protect></protect>
Notes
Links
| Name | URL | Comments |
|---|---|---|
| edit table |
References
See Help:References for how to manage references in EcoliWiki.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Riley, M. et al. (2006) Nucleic Acids Res 34:1-6 (corrected supplemental data from B. Wanner)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 EcoCyc (release 10.6; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 3.0 3.1 EcoCyc (release 11.1; 2007) Keseler, IM et al. (2005) Nucleic Acids Res. 33(Database issue):D334-7
- ↑ 4.0 4.1 EcoGene: Rudd, KE (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60-4.
Categories
- Gene List:REL606
- Gene List:BW2952
- Genes with homologs in Anopheles gambiae
- Genes with homologs in Arabidopsis thaliana
- Genes with homologs in Bos taurus
- Genes with homologs in Caenorhabditis elegans
- Genes with homologs in Ciona intestinalis
- Genes with homologs in Dictyostelium discoideum
- Genes with homologs in Homo sapiens
- Genes with homologs in Mus musculus
- Genes with homologs in Rattus norvegicus
- Genes with homologs in Shigella flexneri
- Genes with homologs in E. coli O157
